bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2005_orf1
Length=124
Score E
Sequences producing significant alignments: (Bits) Value
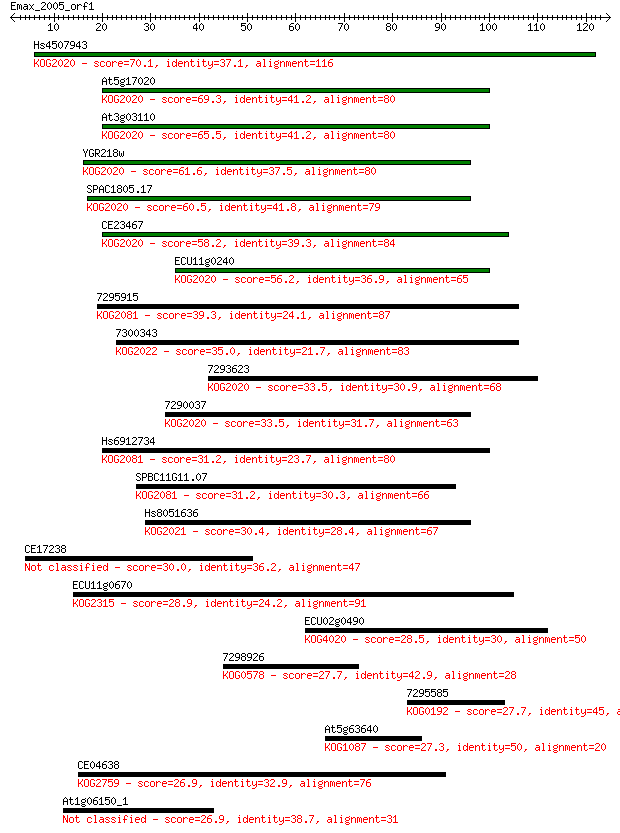
Hs4507943 70.1 1e-12
At5g17020 69.3 2e-12
At3g03110 65.5 2e-11
YGR218w 61.6 3e-10
SPAC1805.17 60.5 7e-10
CE23467 58.2 4e-09
ECU11g0240 56.2 1e-08
7295915 39.3 0.002
7300343 35.0 0.030
7293623 33.5 0.088
7290037 33.5 0.11
Hs6912734 31.2 0.44
SPBC11G11.07 31.2 0.50
Hs8051636 30.4 0.76
CE17238 30.0 1.1
ECU11g0670 28.9 2.8
ECU02g0490 28.5 3.5
7298926 27.7 5.6
7295585 27.7 6.0
At5g63640 27.3 6.3
CE04638 26.9 9.1
At1g06150_1 26.9 9.8
> Hs4507943
Length=1071
Score = 70.1 bits (170), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 43/123 (34%), Positives = 64/123 (52%), Gaps = 11/123 (8%)
Query 6 RALLDPNLPFNEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNV 65
R LLD + + +NLLD VV ++ + +R +A VL K P+AWT V IL
Sbjct 14 RQLLDFSQKLD---INLLDNVVNCLYHGEGAQQR-MAQEVLTHLKEHPDAWTRVDTILEF 69
Query 66 SKDPNTKFFALQILEATITNRWNVLPETERNSTKS-------RSIKNKTCGYKMFIYMQK 118
S++ NTK++ LQILE I RW +LP + K ++ + TC K +Y+ K
Sbjct 70 SQNMNTKYYGLQILENVIKTRWKILPRNQCEGIKKYVVGLIIKTSSDPTCVEKEKVYIGK 129
Query 119 GNI 121
N+
Sbjct 130 LNM 132
> At5g17020
Length=1075
Score = 69.3 bits (168), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 33/80 (41%), Positives = 46/80 (57%), Gaps = 0/80 (0%)
Query 20 VNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFALQIL 79
V +LD VAA F++ ER A ++L + + P+ W V IL + +TKFFALQ+L
Sbjct 15 VGVLDATVAAFFVTGSKEERAAADQILRDLQANPDMWLQVVHILQNTNSLDTKFFALQVL 74
Query 80 EATITNRWNVLPETERNSTK 99
E I RWN LP +R+ K
Sbjct 75 EGVIKYRWNALPVEQRDGMK 94
> At3g03110
Length=1022
Score = 65.5 bits (158), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 33/80 (41%), Positives = 43/80 (53%), Gaps = 0/80 (0%)
Query 20 VNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFALQIL 79
V LLD V A + + ER A +L + K P+ W V IL + +TKFFALQ+L
Sbjct 15 VVLLDATVEAFYSTGSKEERASADNILRDLKANPDTWLQVVHILQNTSSTHTKFFALQVL 74
Query 80 EATITNRWNVLPETERNSTK 99
E I RWN LP +R+ K
Sbjct 75 EGVIKYRWNALPVEQRDGMK 94
> YGR218w
Length=1084
Score = 61.6 bits (148), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 46/80 (57%), Gaps = 1/80 (1%)
Query 16 NEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFA 75
N+ + LLD+VV+ F V++ A +L +F++ P+AW IL S +P +KF A
Sbjct 9 NDLDIALLDQVVST-FYQGSGVQQKQAQEILTKFQDNPDAWQKADQILQFSTNPQSKFIA 67
Query 76 LQILEATITNRWNVLPETER 95
L IL+ IT +W +LP R
Sbjct 68 LSILDKLITRKWKLLPNDHR 87
> SPAC1805.17
Length=966
Score = 60.5 bits (145), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 33/79 (41%), Positives = 46/79 (58%), Gaps = 1/79 (1%)
Query 17 EAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFAL 76
E V LLDRVV F E+ A +VL +F+ P+AW+ IL S+ P TK+ AL
Sbjct 10 ELDVALLDRVVQT-FYQGVGAEQQQAQQVLTQFQAHPDAWSQAYSILEKSEYPQTKYIAL 68
Query 77 QILEATITNRWNVLPETER 95
+L+ IT RW +LP+ +R
Sbjct 69 SVLDKLITTRWKMLPKEQR 87
> CE23467
Length=1080
Score = 58.2 bits (139), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 33/84 (39%), Positives = 48/84 (57%), Gaps = 1/84 (1%)
Query 20 VNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFALQIL 79
V LLD+VV M + E+ A+++L K ++WT V IL S+ +K+FALQIL
Sbjct 24 VTLLDQVVEIMNRMSGK-EQAEANQILMSLKEERDSWTKVDAILQYSQLNESKYFALQIL 82
Query 80 EATITNRWNVLPETERNSTKSRSI 103
E I ++W LP+ +R KS I
Sbjct 83 ETVIQHKWKSLPQVQREGIKSYII 106
> ECU11g0240
Length=1058
Score = 56.2 bits (134), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 42/65 (64%), Gaps = 0/65 (0%)
Query 35 DSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFALQILEATITNRWNVLPETE 94
D+ +++ A R+L +FK +PN+WT V +ILN SK + + ALQ+LE+ I +W++ E
Sbjct 61 DNRKKEEAERILLKFKELPNSWTKVDYILNNSKLQESHYVALQLLESIIKTKWSLFDEGM 120
Query 95 RNSTK 99
+ +
Sbjct 121 KQGLR 125
> 7295915
Length=914
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 21/87 (24%), Positives = 47/87 (54%), Gaps = 0/87 (0%)
Query 19 AVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFALQI 78
+V+++ + ++A+F N+ E++ A++ L +F+ +WT +L+ +D + +FA Q
Sbjct 5 SVDVVYQAISALFQGNNPKEQEKANKWLQDFQKSIYSWTIADELLHQKRDLHANYFAAQT 64
Query 79 LEATITNRWNVLPETERNSTKSRSIKN 105
+ I N ++ LP S + I +
Sbjct 65 MRNKIQNSFSELPPHTHESLRDSLITH 91
> 7300343
Length=971
Score = 35.0 bits (79), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 18/83 (21%), Positives = 37/83 (44%), Gaps = 1/83 (1%)
Query 23 LDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFALQILEAT 82
L+ V + + SN S + + H L + + P AW ++ + K +FF L +
Sbjct 9 LEEAVVSFYRSN-SQNQAITHEWLTDAEASPQAWQFSWQLMQLGKSQEVQFFGAITLHSK 67
Query 83 ITNRWNVLPETERNSTKSRSIKN 105
+ W+ +P R K + +++
Sbjct 68 LMKHWHEVPPENREELKQKILES 90
> 7293623
Length=1241
Score = 33.5 bits (75), Expect = 0.088, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 28/70 (40%), Gaps = 2/70 (2%)
Query 42 AHRVLGEFKNMPNAWTHVAFILNVSKDPN--TKFFALQILEATITNRWNVLPETERNSTK 99
A+ FK V L S N + F LQ++E TI RWN + E+ K
Sbjct 34 AYMACERFKEESPMCAQVGLFLASSPQSNQQVRHFGLQLIEYTIKYRWNCITHEEKVYIK 93
Query 100 SRSIKNKTCG 109
+IK G
Sbjct 94 DNAIKMLNVG 103
> 7290037
Length=186
Score = 33.5 bits (75), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 36/66 (54%), Gaps = 4/66 (6%)
Query 33 SNDSVERDVAHRVLGEFKNMPNAWT---HVAFILNVSKDPNTKFFALQILEATITNRWNV 89
++++ +R++ +L FK+ P AW VA + +++ FF+ LE TIT RW
Sbjct 21 TSNARKREIETNLLA-FKSQPEAWQLCLRVATSSDTTENQFLWFFSTSTLEHTITRRWTQ 79
Query 90 LPETER 95
L T++
Sbjct 80 LTSTDK 85
> Hs6912734
Length=923
Score = 31.2 bits (69), Expect = 0.44, Method: Composition-based stats.
Identities = 19/80 (23%), Positives = 37/80 (46%), Gaps = 0/80 (0%)
Query 20 VNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFALQIL 79
+ L+ + V A++ D ++ A LGE + +AW +L + +D + +FA Q +
Sbjct 8 LQLVYQAVQALYHDPDPSGKERASFWLGELQRSVHAWEISDQLLQIRQDVESCYFAAQTM 67
Query 80 EATITNRWNVLPETERNSTK 99
+ I + LP S +
Sbjct 68 KMKIQTSFYELPTDSHASLR 87
> SPBC11G11.07
Length=378
Score = 31.2 bits (69), Expect = 0.50, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 31/67 (46%), Gaps = 1/67 (1%)
Query 27 VAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDP-NTKFFALQILEATITN 85
+A ++ + D ++ A+ L EF+ P AW ILN K FA Q L I
Sbjct 8 LATLYANTDREQKLQANNYLEEFQKSPAAWQICFSILNQDDSSIEAKLFAAQTLRQKIVY 67
Query 86 RWNVLPE 92
++ LP+
Sbjct 68 DFHQLPK 74
> Hs8051636
Length=962
Score = 30.4 bits (67), Expect = 0.76, Method: Composition-based stats.
Identities = 19/72 (26%), Positives = 33/72 (45%), Gaps = 5/72 (6%)
Query 29 AMFLSNDSVERDVAHRVLGEFKNM---PNAWTHVAFIL--NVSKDPNTKFFALQILEATI 83
A+ N + + D R L F+ + P+AW A L D + KFF Q+LE +
Sbjct 5 ALLGLNPNADSDFRQRALAYFEQLKISPDAWQVCAEALAQRTYSDDHVKFFCFQVLEHQV 64
Query 84 TNRWNVLPETER 95
+++ L ++
Sbjct 65 KYKYSELTTVQQ 76
> CE17238
Length=1028
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 23/47 (48%), Gaps = 0/47 (0%)
Query 4 DPRALLDPNLPFNEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFK 50
D R LL N A ++ D+ V+A+F DSV D +VL K
Sbjct 79 DLRGLLKNKYTVNSFAADVQDKTVSAVFSLRDSVSADDVQKVLAGSK 125
> ECU11g0670
Length=496
Score = 28.9 bits (63), Expect = 2.8, Method: Composition-based stats.
Identities = 22/95 (23%), Positives = 45/95 (47%), Gaps = 4/95 (4%)
Query 14 PFNEAAVNLLDRVVAA--MFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFI--LNVSKDP 69
PF+ + V++L+ ++ +FL+ +E+D HRVL KN + + ++
Sbjct 161 PFHNSRVSMLEFSISGEFVFLATKKLEKDGMHRVLRIGKNTVEVLHSLKELQGFSLKTHV 220
Query 70 NTKFFALQILEATITNRWNVLPETERNSTKSRSIK 104
+ K+ L ++ + + N + E T+ RS K
Sbjct 221 SNKYTLLLLMTSYVNNSYFAESELYLYDTEKRSFK 255
> ECU02g0490
Length=112
Score = 28.5 bits (62), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 15/50 (30%), Positives = 25/50 (50%), Gaps = 5/50 (10%)
Query 62 ILNVSKDPNTKFFALQILEATITNRWNVLPETERNSTKSRSIKNKTCGYK 111
++ S DP TK + + +T+ + +ER K R+ K+ TCG K
Sbjct 14 MMRKSTDPRTK-----MQDEYLTDEDKAIQRSERPPAKKRACKDCTCGLK 58
> 7298926
Length=704
Score = 27.7 bits (60), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 12/28 (42%), Positives = 14/28 (50%), Gaps = 0/28 (0%)
Query 45 VLGEFKNMPNAWTHVAFILNVSKDPNTK 72
V GEF MP AW + N+SK K
Sbjct 100 VTGEFTGMPEAWARLLMNSNISKQEQKK 127
> 7295585
Length=678
Score = 27.7 bits (60), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 9/20 (45%), Positives = 15/20 (75%), Gaps = 0/20 (0%)
Query 83 ITNRWNVLPETERNSTKSRS 102
+TNRW+ +PE E N +++ S
Sbjct 371 VTNRWDAIPEEESNESRNDS 390
> At5g63640
Length=447
Score = 27.3 bits (59), Expect = 6.3, Method: Composition-based stats.
Identities = 10/20 (50%), Positives = 16/20 (80%), Gaps = 0/20 (0%)
Query 66 SKDPNTKFFALQILEATITN 85
SK+PNT+ +A+Q+LE + N
Sbjct 49 SKNPNTQLYAVQLLEMLMNN 68
> CE04638
Length=451
Score = 26.9 bits (58), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 25/85 (29%), Positives = 36/85 (42%), Gaps = 9/85 (10%)
Query 15 FNEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEF-KNMPNAWTHVAFI--------LNV 65
N+ LL +VA + SND + VA +GEF + P V + L
Sbjct 363 LNDNRQELLKLLVAMLEKSNDPLVLCVAAHDIGEFVRYYPRGKLKVEQLGGKEAMMRLLT 422
Query 66 SKDPNTKFFALQILEATITNRWNVL 90
KDPN ++ AL + + N W L
Sbjct 423 VKDPNVRYHALLAAQKLMINNWKDL 447
> At1g06150_1
Length=789
Score = 26.9 bits (58), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 12 NLPFNEAAVNLLDRVVAAMFLSNDSVERDVA 42
+L F ++ NLLD VVA+M + +V R+++
Sbjct 401 HLTFESSSENLLDAVVASMSNGDGNVRREIS 431
Lambda K H
0.320 0.132 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187579072
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40