bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2004_orf1
Length=152
Score E
Sequences producing significant alignments: (Bits) Value
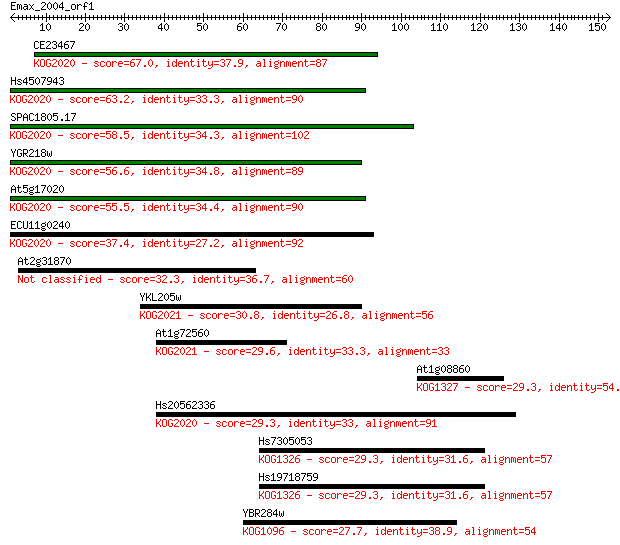
CE23467 67.0 1e-11
Hs4507943 63.2 2e-10
SPAC1805.17 58.5 5e-09
YGR218w 56.6 2e-08
At5g17020 55.5 4e-08
ECU11g0240 37.4 0.012
At2g31870 32.3 0.34
YKL205w 30.8 1.1
At1g72560 29.6 2.2
At1g08860 29.3 3.2
Hs20562336 29.3 3.2
Hs7305053 29.3 3.2
Hs19718759 29.3 3.3
YBR284w 27.7 8.9
> CE23467
Length=1080
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 33/87 (37%), Positives = 55/87 (63%), Gaps = 7/87 (8%)
Query 7 QFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFETELLPLLLQY 66
QF+E++ +C+ +L+ S+ +++Q TLK L FL WIP+G+VFET + LL +
Sbjct 200 QFQEVFTLCVSILEKCPSN-------SMVQATLKTLQRFLTWIPVGYVFETNITELLSEN 252
Query 67 FYEDVSFRIDCLRCLTEVSSLQLNINE 93
F +R+ L+CLTE+S +Q+ N+
Sbjct 253 FLSLEVYRVIALQCLTEISQIQVETND 279
> Hs4507943
Length=1071
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 55/90 (61%), Gaps = 7/90 (7%)
Query 1 RDELTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFETELL 60
+D + +F +++ +C FV+++ ++P L+ TL+ L FL WIPLG++FET+L+
Sbjct 195 KDSMCNEFSQIFQLCQFVMENSQNAP-------LVHATLETLLRFLNWIPLGYIFETKLI 247
Query 61 PLLLQYFYEDVSFRIDCLRCLTEVSSLQLN 90
L+ F FR L+CLTE++ + ++
Sbjct 248 STLIYKFLNVPMFRNVSLKCLTEIAGVSVS 277
> SPAC1805.17
Length=966
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 35/106 (33%), Positives = 58/106 (54%), Gaps = 11/106 (10%)
Query 1 RDELTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFETELL 60
++++ +F E++ +C +L+ K +LI+ TL L FL WIPLG++FET ++
Sbjct 183 KNQMCGEFAEIFQLCSQILER-------AQKPSLIKATLGTLLRFLNWIPLGYIFETNIV 235
Query 61 PLLLQYFYEDVSFRIDCLRCLTEVSSL----QLNINELNLFSQQMT 102
L+ F FR + CLTE++SL Q N + +F+ MT
Sbjct 236 ELITNRFLNVPDFRNVTIECLTEIASLTSQPQYNDKFVTMFNLVMT 281
> YGR218w
Length=1084
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/89 (34%), Positives = 53/89 (59%), Gaps = 7/89 (7%)
Query 1 RDELTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFETELL 60
++ ++K+F++++ +C VL+ SS +LI TL+ L +L WIP +++ET +L
Sbjct 183 KNSMSKEFEQIFKLCFQVLEQGSSS-------SLIVATLESLLRYLHWIPYRYIYETNIL 235
Query 61 PLLLQYFYEDVSFRIDCLRCLTEVSSLQL 89
LL F R L+CLTEVS+L++
Sbjct 236 ELLSTKFMTSPDTRAITLKCLTEVSNLKI 264
> At5g17020
Length=1075
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 31/90 (34%), Positives = 53/90 (58%), Gaps = 8/90 (8%)
Query 1 RDELTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFETELL 60
+ L +FK ++ +C++VL + + LI+ TL L +L WIPLG++FE+ LL
Sbjct 186 KQSLNSEFKLIHELCLYVLSA-------SQRQDLIRATLSALHAYLSWIPLGYIFESTLL 238
Query 61 PLLLQYFYEDVSFRIDCLRCLTEVSSLQLN 90
LL+ F+ ++R ++CLTEV++L
Sbjct 239 ETLLK-FFPVPAYRNLTIQCLTEVAALNFG 267
> ECU11g0240
Length=1058
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 50/92 (54%), Gaps = 5/92 (5%)
Query 1 RDELTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFETELL 60
R++L +F +++ +L+ S + + +L++ TL+ F + +PL F+F TE++
Sbjct 215 RNQLKIEFPQIFGFIKTILEY---SRNTEMDESLLEATLESFQCFCESMPLDFIFLTEII 271
Query 61 PLLLQYFYEDVSFRIDCLRCLTEVSSLQLNIN 92
L+L++ + + CL CL E+ L N N
Sbjct 272 DLVLEHL--NSMHSVACLSCLIEIVDLGRNRN 301
> At2g31870
Length=997
Score = 32.3 bits (72), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 32/65 (49%), Gaps = 9/65 (13%)
Query 3 ELTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFL----KWIPLGFV-FET 57
E+ + K L + L SF H T + +KCL H+ +W+P GFV FE
Sbjct 680 EVDRSLKNLQGINFSGLFSFPYMRH----CTKQENKIKCLIHYFGRICRWMPTGFVSFER 735
Query 58 ELLPL 62
++LPL
Sbjct 736 KILPL 740
> YKL205w
Length=1100
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 15/57 (26%), Positives = 28/57 (49%), Gaps = 1/57 (1%)
Query 34 LIQQTLKCLAHFLKWIPLGFVFETELLPLLLQYFYEDVS-FRIDCLRCLTEVSSLQL 89
LI TL C+ F+ WI + + + L L Y + ++ +I C C+ + S ++
Sbjct 226 LINSTLDCIGSFISWIDINLIIDANNYYLQLIYKFLNLKETKISCYNCILAIISKKM 282
> At1g72560
Length=993
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 11/33 (33%), Positives = 16/33 (48%), Gaps = 0/33 (0%)
Query 38 TLKCLAHFLKWIPLGFVFETELLPLLLQYFYED 70
L C+ F+ WI +G V +PLL + D
Sbjct 213 VLDCMRRFVSWIDIGLVANDAFVPLLFELILSD 245
> At1g08860
Length=644
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 12/22 (54%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 104 LWAQLSDKAAHTPKQSLQYNDP 125
L++ + DKAAHT QSL N P
Sbjct 509 LFSNVVDKAAHTASQSLSQNSP 530
> Hs20562336
Length=1065
Score = 29.3 bits (64), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 30/120 (25%), Positives = 44/120 (36%), Gaps = 29/120 (24%)
Query 38 TLKCLAHFLKWIPLGFVFETELLPLLLQY--FYEDVSFR--------------------- 74
L+CLAH WIPL LL + + F D+ R
Sbjct 189 ALECLAHLFSWIPLSASITPSLLTTIFHFARFGCDIRARKMASVNGSSQNCVSGQERGRL 248
Query 75 -IDCLRCLTEVSS---LQLNINE--LNLFSQQMTLLWAQLSDKAAHTPKQSLQYNDPTSV 128
+ + C+ E+ S + + E L +F Q LL D AHT K L+ D + +
Sbjct 249 GVLAMSCINELMSKNCVPMEFEEYLLRMFQQTFYLLQKITKDNNAHTVKSRLEELDESYI 308
> Hs7305053
Length=2061
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 32/64 (50%), Gaps = 7/64 (10%)
Query 64 LQYFYEDVSFRIDCLRCLTEVSS-LQLNINEL------NLFSQQMTLLWAQLSDKAAHTP 116
L ++ED+S R+D + L ++ LQ NI L + + Q+ LW +L D+
Sbjct 643 LTSYWEDISHRLDAVNTLLAMAERLQTNIEALKSGIQGKIPANQLAELWLKLIDEVIEDT 702
Query 117 KQSL 120
+ +L
Sbjct 703 RYTL 706
> Hs19718759
Length=2048
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 32/64 (50%), Gaps = 7/64 (10%)
Query 64 LQYFYEDVSFRIDCLRCLTEVSS-LQLNINEL------NLFSQQMTLLWAQLSDKAAHTP 116
L ++ED+S R+D + L ++ LQ NI L + + Q+ LW +L D+
Sbjct 630 LTSYWEDISHRLDAVNTLLAMAERLQTNIEALKSGIQGKIPANQLAELWLKLIDEVIEDT 689
Query 117 KQSL 120
+ +L
Sbjct 690 RYTL 693
> YBR284w
Length=797
Score = 27.7 bits (60), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 28/54 (51%), Gaps = 4/54 (7%)
Query 60 LPLLLQYFYEDVSFRIDCLRCLTEVSSLQLNINELNLFSQQMTLLWAQLSDKAA 113
L L YF+E FRID +CL E+S L+ N + S+Q L QL + A
Sbjct 119 LAHLPSYFFEQTHFRID-RKCLLEMSKLRRNYLTI---SKQDALSCPQLHSRVA 168
Lambda K H
0.327 0.140 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1925034518
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40