bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1968_orf1
Length=85
Score E
Sequences producing significant alignments: (Bits) Value
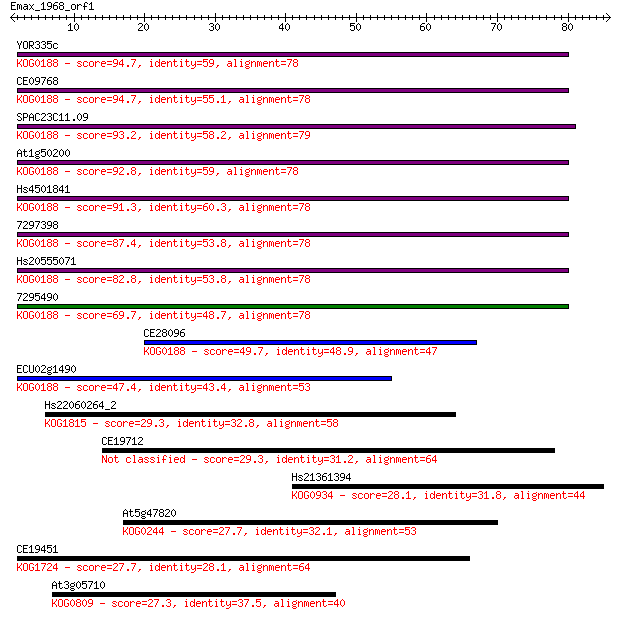
YOR335c 94.7 3e-20
CE09768 94.7 4e-20
SPAC23C11.09 93.2 1e-19
At1g50200 92.8 1e-19
Hs4501841 91.3 4e-19
7297398 87.4 5e-18
Hs20555071 82.8 1e-16
7295490 69.7 1e-12
CE28096 49.7 1e-06
ECU02g1490 47.4 6e-06
Hs22060264_2 29.3 2.1
CE19712 29.3 2.1
Hs21361394 28.1 4.1
At5g47820 27.7 4.8
CE19451 27.7 4.9
At3g05710 27.3 6.2
> YOR335c
Length=958
Score = 94.7 bits (234), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 46/78 (58%), Positives = 55/78 (70%), Gaps = 1/78 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
+QFNRE +L PLPA +DTG ERLVSVLQ+ +SNYDTD+F P+FERI + P
Sbjct 224 IQFNREQDGSLKPLPAKHIDTGMGFERLVSVLQDVRSNYDTDVFTPLFERIQEITSVRP- 282
Query 62 YQGKVGEEDKNKIDTAYR 79
Y G GE DK+ IDTAYR
Sbjct 283 YSGNFGENDKDGIDTAYR 300
> CE09768
Length=968
Score = 94.7 bits (234), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 43/78 (55%), Positives = 56/78 (71%), Gaps = 1/78 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
+QFNRE L PLPA +D G LERL++V+Q+K SNYDTD+FQPIFE IH + +
Sbjct 220 IQFNREEGGVLKPLPAKHIDCGLGLERLIAVMQDKTSNYDTDIFQPIFEAIHK-GSGVRA 278
Query 62 YQGKVGEEDKNKIDTAYR 79
Y G +G+EDK+ +D AYR
Sbjct 279 YTGFIGDEDKDGVDMAYR 296
> SPAC23C11.09
Length=959
Score = 93.2 bits (230), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 46/79 (58%), Positives = 54/79 (68%), Gaps = 1/79 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
+QFNRE +L PLP VDTG ERLVSV+QNK SNYDTD+F PIF +I P
Sbjct 225 IQFNREKDGSLRPLPNRHVDTGMGFERLVSVIQNKTSNYDTDVFSPIFAKIQELTNARP- 283
Query 62 YQGKVGEEDKNKIDTAYRA 80
Y GK+G+ED + IDTAYR
Sbjct 284 YTGKMGDEDVDGIDTAYRV 302
> At1g50200
Length=1027
Score = 92.8 bits (229), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 46/78 (58%), Positives = 54/78 (69%), Gaps = 1/78 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
+QFNRE+ +L PLPA VDTG ERL SVLQNK SNYDTD+F PIF+ I P
Sbjct 273 IQFNRESDGSLKPLPAKHVDTGMGFERLTSVLQNKMSNYDTDVFMPIFDDIQKATGARP- 331
Query 62 YQGKVGEEDKNKIDTAYR 79
Y GKVG ED +++D AYR
Sbjct 332 YSGKVGPEDVDRVDMAYR 349
> Hs4501841
Length=968
Score = 91.3 bits (225), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 47/78 (60%), Positives = 51/78 (65%), Gaps = 1/78 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
+Q+NRE L PLP +DTG LERLVSVLQNK SNYDTDLF P FE I P
Sbjct 220 IQYNREADGILKPLPKKSIDTGMGLERLVSVLQNKMSNYDTDLFVPYFEAIQKGTGARP- 278
Query 62 YQGKVGEEDKNKIDTAYR 79
Y GKVG ED + ID AYR
Sbjct 279 YTGKVGAEDADGIDMAYR 296
> 7297398
Length=966
Score = 87.4 bits (215), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 42/78 (53%), Positives = 55/78 (70%), Gaps = 1/78 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
+Q+NRE+ +L LP +D G ERLVSV+QNK+SNYDTDLF P+F+ I + T P
Sbjct 219 IQYNRESDGSLKQLPKKHIDCGMGFERLVSVIQNKRSNYDTDLFVPLFDAIQA-GTGAPP 277
Query 62 YQGKVGEEDKNKIDTAYR 79
YQG+VG +D + ID AYR
Sbjct 278 YQGRVGADDVDGIDMAYR 295
> Hs20555071
Length=985
Score = 82.8 bits (203), Expect = 1e-16, Method: Composition-based stats.
Identities = 42/78 (53%), Positives = 49/78 (62%), Gaps = 1/78 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
MQ NRE +L PLP VDTG LERLV+VLQ K S YDTDLF P+ I + P
Sbjct 246 MQHNREADGSLQPLPQRHVDTGMGLERLVAVLQGKHSTYDTDLFSPLLNAIQQ-GCRAPP 304
Query 62 YQGKVGEEDKNKIDTAYR 79
Y G+VG D+ + DTAYR
Sbjct 305 YLGRVGVADEGRTDTAYR 322
> 7295490
Length=1012
Score = 69.7 bits (169), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 38/79 (48%), Positives = 48/79 (60%), Gaps = 2/79 (2%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
+Q+NR +++ LPA VDTG ERL +VLQNK SNYDTDLF PIF+ I K P
Sbjct 227 IQYNRHADGSISQLPAHHVDTGMGFERLTAVLQNKSSNYDTDLFTPIFDGIQQ-AAKTPV 285
Query 62 YQGKVGEEDKNKI-DTAYR 79
Y G + + DT+YR
Sbjct 286 YSGSFPDGGNAAVLDTSYR 304
> CE28096
Length=793
Score = 49.7 bits (117), Expect = 1e-06, Method: Composition-based stats.
Identities = 23/47 (48%), Positives = 31/47 (65%), Gaps = 2/47 (4%)
Query 20 VDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPKYQGKV 66
+DTG ERL+SV+QNK SN+DTD+F PI E+ K +Y G +
Sbjct 248 IDTGMGFERLLSVVQNKTSNFDTDVFTPILEKTSELAKK--QYTGSL 292
> ECU02g1490
Length=864
Score = 47.4 bits (111), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 23/53 (43%), Positives = 33/53 (62%), Gaps = 1/53 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHS 54
+++NR + L PL C+DTG LERL+S+L + KSNY D F I + + S
Sbjct 204 IEYNRTTS-GLAPLERKCIDTGIGLERLLSILMDVKSNYLIDSFSTIIKAMES 255
> Hs22060264_2
Length=526
Score = 29.3 bits (64), Expect = 2.1, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 30/63 (47%), Gaps = 5/63 (7%)
Query 6 RENAQTLTPLPAPCVDTGTVL-----ERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLP 60
R+ A +L L A C+ TG L ERL+++LQ+ ++ L P E + +P
Sbjct 400 RDLASSLRLLRADCLSTGMELLRRIQERLLAILQHSAQDFRVGLQSPSVEAWEAKGPNMP 459
Query 61 KYQ 63
Q
Sbjct 460 GSQ 462
> CE19712
Length=194
Score = 29.3 bits (64), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 20/67 (29%), Positives = 31/67 (46%), Gaps = 6/67 (8%)
Query 14 PLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFER---IHSFNTKLPKYQGKVGEED 70
P PAP T TV E + ++ QN D FQ +F+ + + K ++ K G D
Sbjct 19 PTPAP---TLTVAEEMEAIFQNLTEEAQNDYFQILFDHTLTLAELDQKWDEWAEKHGIAD 75
Query 71 KNKIDTA 77
K + T+
Sbjct 76 KWAVYTS 82
> Hs21361394
Length=159
Score = 28.1 bits (61), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 14/44 (31%), Positives = 24/44 (54%), Gaps = 0/44 (0%)
Query 41 DTDLFQPIFERIHSFNTKLPKYQGKVGEEDKNKIDTAYRAGGQV 84
DT+ I+E IH+F L +Y +V E D + +T + + Q+
Sbjct 73 DTENEMAIYEFIHNFVEVLDEYFSRVSELDVSFFNTVFHSTWQM 116
> At5g47820
Length=1049
Score = 27.7 bits (60), Expect = 4.8, Method: Composition-based stats.
Identities = 17/56 (30%), Positives = 26/56 (46%), Gaps = 3/56 (5%)
Query 17 APCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK---YQGKVGEE 69
A C+D G+V S N +S+ +F E++ NT P+ Y G + EE
Sbjct 206 AACLDQGSVSRATGSTNMNNQSSRSHAIFTITVEQMRKINTDSPENGAYNGSLKEE 261
> CE19451
Length=184
Score = 27.7 bits (60), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 18/64 (28%), Positives = 34/64 (53%), Gaps = 8/64 (12%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
+ ++ ENA+++ P+P V+ G L+ +V ++ K++ P+ E S NT LP
Sbjct 51 LGYSAENAESMVPIPIEKVN-GKTLKLVVEWCEHHKAD-------PVPEAYPSGNTVLPV 102
Query 62 YQGK 65
+ K
Sbjct 103 WDRK 106
> At3g05710
Length=331
Score = 27.3 bits (59), Expect = 6.2, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 7 ENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQ 46
E AQ + L A +D GT+++R+ +QN S D L Q
Sbjct 254 ELAQIMKDLSALVIDQGTIVDRIDYNIQNVASTVDDGLKQ 293
Lambda K H
0.317 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194132014
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40