bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1916_orf1
Length=156
Score E
Sequences producing significant alignments: (Bits) Value
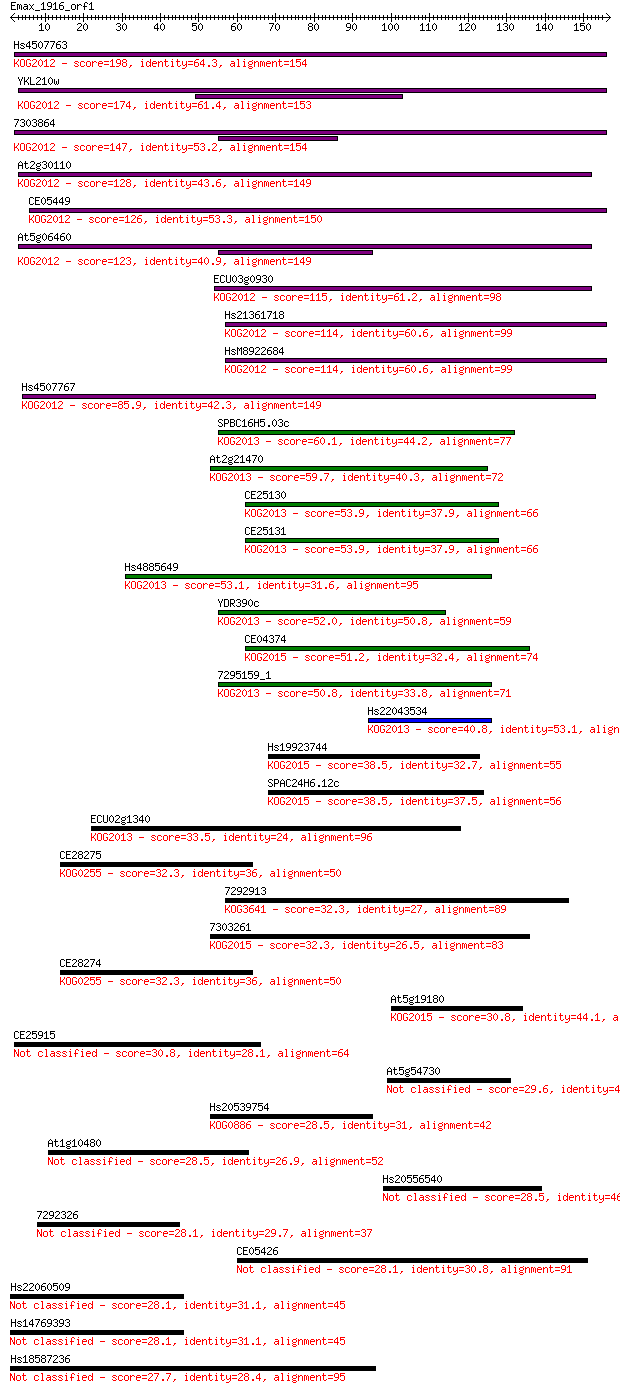
Hs4507763 198 3e-51
YKL210w 174 6e-44
7303864 147 6e-36
At2g30110 128 4e-30
CE05449 126 1e-29
At5g06460 123 2e-28
ECU03g0930 115 3e-26
Hs21361718 114 6e-26
HsM8922684 114 7e-26
Hs4507767 85.9 3e-17
SPBC16H5.03c 60.1 2e-09
At2g21470 59.7 2e-09
CE25130 53.9 1e-07
CE25131 53.9 1e-07
Hs4885649 53.1 2e-07
YDR390c 52.0 5e-07
CE04374 51.2 7e-07
7295159_1 50.8 1e-06
Hs22043534 40.8 0.001
Hs19923744 38.5 0.005
SPAC24H6.12c 38.5 0.005
ECU02g1340 33.5 0.15
CE28275 32.3 0.34
7292913 32.3 0.34
7303261 32.3 0.34
CE28274 32.3 0.35
At5g19180 30.8 0.98
CE25915 30.8 1.1
At5g54730 29.6 2.5
Hs20539754 28.5 5.1
At1g10480 28.5 5.4
Hs20556540 28.5 5.8
7292326 28.1 6.5
CE05426 28.1 6.7
Hs22060509 28.1 7.6
Hs14769393 28.1 7.8
Hs18587236 27.7 8.5
> Hs4507763
Length=1058
Score = 198 bits (504), Expect = 3e-51, Method: Composition-based stats.
Identities = 99/154 (64%), Positives = 122/154 (79%), Gaps = 4/154 (2%)
Query 2 EIPVFTPKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPIEFEK 61
++P FTPK +KI V+D EL S N++V ++ + L+ LPS +L G K+ PI+FEK
Sbjct 795 QVPEFTPKSGVKIHVSDQELQSANASVD---DSRLEELKATLPSPDKLPGFKMYPIDFEK 851
Query 62 DDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYK 121
DDD+NFH+DFIVAASNLRAENY+IP ADRHKSKLIAG+IIPAIATTTA V GLV +ELYK
Sbjct 852 DDDSNFHMDFIVAASNLRAENYDIPSADRHKSKLIAGKIIPAIATTTAAVVGLVCLELYK 911
Query 122 LVQGFTNTLEPYKNGFVNLALPFFGFSTPIAVPK 155
+VQG L+ YKNGF+NLALPFFGFS P+A P+
Sbjct 912 VVQGH-RQLDSYKNGFLNLALPFFGFSEPLAAPR 944
> YKL210w
Length=1024
Score = 174 bits (441), Expect = 6e-44, Method: Compositional matrix adjust.
Identities = 94/153 (61%), Positives = 110/153 (71%), Gaps = 3/153 (1%)
Query 3 IPVFTPKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPIEFEKD 62
IP FTP NLKI VND + + N+N + L LP S LAG K+ P++FEKD
Sbjct 767 IPEFTPNANLKIQVNDDDPDP-NANAANG-SDEIDQLVSSLPDPSTLAGFKLEPVDFEKD 824
Query 63 DDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYKL 122
DDTN HI+FI A SN RA+NY I ADR K+K IAGRIIPAIATTT+LV+GLV +ELYKL
Sbjct 825 DDTNHHIEFITACSNCRAQNYFIETADRQKTKFIAGRIIPAIATTTSLVTGLVNLELYKL 884
Query 123 VQGFTNTLEPYKNGFVNLALPFFGFSTPIAVPK 155
+ T+ +E YKNGFVNLALPFFGFS PIA PK
Sbjct 885 IDNKTD-IEQYKNGFVNLALPFFGFSEPIASPK 916
Score = 33.1 bits (74), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 24/62 (38%), Positives = 30/62 (48%), Gaps = 9/62 (14%)
Query 49 LAGVKINPIEFEKDDDTNFHIDFIVAASNLRAENYEI--------PPADRHKSKLIAGRI 100
+G K P E D N H F+VA ++LRA NY I P D +KS +I I
Sbjct 708 WSGAKRAPTPLEFDIYNNDHFHFVVAGASLRAYNYGIKSDDSNSKPNVDEYKS-VIDHMI 766
Query 101 IP 102
IP
Sbjct 767 IP 768
> 7303864
Length=1008
Score = 147 bits (372), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 82/166 (49%), Positives = 104/166 (62%), Gaps = 24/166 (14%)
Query 2 EIPVFTPKQNLKIAVN------------DSELNSENSNVPTDLNTHFKNLQKQLPSVSEL 49
++P F P+ +KI N D EL+ + V ++ KN K
Sbjct 747 KVPEFKPRSGVKIETNEAAAAASANNFDDGELDQDR--VDKIISELLKNADKS------- 797
Query 50 AGVKINPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTA 109
KI P+EFEKDDD+N H+DFIVA SNLRA NY+IPPADRHKSKLIAG+IIPAIATTT+
Sbjct 798 --SKITPLEFEKDDDSNLHMDFIVACSNLRAANYKIPPADRHKSKLIAGKIIPAIATTTS 855
Query 110 LVSGLVFIELYKLVQGFTNTLEPYKNGFVNLALPFFGFSTPIAVPK 155
++SGL +E+ KL+ G + L +KNGF NLALPF FS P+ K
Sbjct 856 VLSGLAVLEVIKLIVGHRD-LVKFKNGFANLALPFMAFSEPLPAAK 900
Score = 32.0 bits (71), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 16/31 (51%), Positives = 22/31 (70%), Gaps = 2/31 (6%)
Query 55 NPIEFEKDDDTNFHIDFIVAASNLRAENYEI 85
+P+ F+ +D H+DFI AA+NLRAE Y I
Sbjct 702 DPLVFDVNDP--MHLDFIYAAANLRAEVYGI 730
> At2g30110
Length=1080
Score = 128 bits (322), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 65/154 (42%), Positives = 96/154 (62%), Gaps = 11/154 (7%)
Query 3 IPVFTPKQNLKIAVND-----SELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPI 57
+P F P+Q+ KI ++ + + +++ V DL + L ++ PI
Sbjct 824 VPDFEPRQDAKIVTDEKATTLTTASVDDAAVIDDLIAKIDQCRHNLS-----PDFRMKPI 878
Query 58 EFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFI 117
+FEKDDDTN+H+D I +N+RA NY IP D+ K+K IAGRIIPAIAT+TA+ +GLV +
Sbjct 879 QFEKDDDTNYHMDVIAGLANMRARNYSIPEVDKLKAKFIAGRIIPAIATSTAMATGLVCL 938
Query 118 ELYKLVQGFTNTLEPYKNGFVNLALPFFGFSTPI 151
ELYK++ G + +E Y+N F NLALP F + P+
Sbjct 939 ELYKVLDG-GHKVEAYRNTFANLALPLFSMAEPL 971
> CE05449
Length=1113
Score = 126 bits (317), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 80/157 (50%), Positives = 111/157 (70%), Gaps = 7/157 (4%)
Query 6 FTPKQNLKIAVNDSELNSEN----SNVPTDLNTHFKNLQKQLPSVSELAGVKINPIEFEK 61
F PK +KIAV D+E +N S++ D + + L+ +L +++ + K+N ++FEK
Sbjct 849 FEPKSGVKIAVTDAEAKEQNERGASSMIVDDDAAIEALKLKLATLNVKSTSKLNCVDFEK 908
Query 62 DDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYK 121
DDD+N H++FI AASNLRAENY+I PADR ++K IAG+IIPAIATTTA V+GLV IELYK
Sbjct 909 DDDSNHHMEFITAASNLRAENYDILPADRMRTKQIAGKIIPAIATTTAAVAGLVCIELYK 968
Query 122 LVQ--GFTNT-LEPYKNGFVNLALPFFGFSTPIAVPK 155
+V G T +E +KN F+NL++PFF + PI PK
Sbjct 969 VVDANGIPKTPMERFKNTFLNLSMPFFSSAEPIGAPK 1005
> At5g06460
Length=1077
Score = 123 bits (308), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 61/154 (39%), Positives = 95/154 (61%), Gaps = 11/154 (7%)
Query 3 IPVFTPKQNLKIAVND-----SELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPI 57
+P F PK++ I ++ S + +++ V +LN + L ++ I
Sbjct 821 VPDFEPKKDATIVTDEKATTLSTASVDDAAVIDELNAKLVRCRMSLQP-----EFRMKAI 875
Query 58 EFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFI 117
+FEKDDDTN+H+D I +N+RA NY +P D+ K+K IAGRIIPAIAT+TA+ +G V +
Sbjct 876 QFEKDDDTNYHMDMIAGLANMRARNYSVPEVDKLKAKFIAGRIIPAIATSTAMATGFVCL 935
Query 118 ELYKLVQGFTNTLEPYKNGFVNLALPFFGFSTPI 151
E+YK++ G ++ +E Y+N F NLALP F + P+
Sbjct 936 EMYKVLDG-SHKVEDYRNTFANLALPLFSMAEPV 968
Score = 32.3 bits (72), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 26/40 (65%), Gaps = 2/40 (5%)
Query 55 NPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSK 94
P++F D + HI+F++AAS LRAE + IP + K++
Sbjct 772 RPLQFSSTDLS--HINFVMAASILRAETFGIPTPEWAKTR 809
> ECU03g0930
Length=991
Score = 115 bits (288), Expect = 3e-26, Method: Composition-based stats.
Identities = 60/98 (61%), Positives = 71/98 (72%), Gaps = 2/98 (2%)
Query 54 INPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSG 113
I P FEKDDDTNFH+DF+ AA+NLRA NY+I ADR K IAGRIIPAIATTTA+VSG
Sbjct 792 IVPCIFEKDDDTNFHVDFLYAAANLRAINYKIKQADRLTVKGIAGRIIPAIATTTAVVSG 851
Query 114 LVFIELYKLVQGFTNTLEPYKNGFVNLALPFFGFSTPI 151
L +E+ K G T +KN F+NLALPFF + P+
Sbjct 852 LAVLEMIKYALGVDYT--KHKNSFLNLALPFFASTDPV 887
> Hs21361718
Length=1052
Score = 114 bits (286), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 60/99 (60%), Positives = 71/99 (71%), Gaps = 2/99 (2%)
Query 57 IEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVF 116
+ FEKDDD N HIDFI AASNLRA+ Y I PADR K+K IAG+IIPAIATTTA VSGLV
Sbjct 847 LSFEKDDDHNGHIDFITAASNLRAKMYSIEPADRFKTKRIAGKIIPAIATTTATVSGLVA 906
Query 117 IELYKLVQGFTNTLEPYKNGFVNLALPFFGFSTPIAVPK 155
+E+ K+ G+ E YKN F+NLA+P F+ V K
Sbjct 907 LEMIKVTGGY--PFEVYKNCFLNLAIPIVVFTETTEVRK 943
> HsM8922684
Length=459
Score = 114 bits (285), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 60/99 (60%), Positives = 71/99 (71%), Gaps = 2/99 (2%)
Query 57 IEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVF 116
+ FEKDDD N HIDFI AASNLRA+ Y I PADR K+K IAG+IIPAIATTTA VSGLV
Sbjct 254 LSFEKDDDHNGHIDFITAASNLRAKMYSIEPADRFKTKRIAGKIIPAIATTTATVSGLVA 313
Query 117 IELYKLVQGFTNTLEPYKNGFVNLALPFFGFSTPIAVPK 155
+E+ K+ G+ E YKN F+NLA+P F+ V K
Sbjct 314 LEMIKVTGGY--PFEAYKNCFLNLAIPIVVFTETTEVRK 350
> Hs4507767
Length=1011
Score = 85.9 bits (211), Expect = 3e-17, Method: Composition-based stats.
Identities = 63/149 (42%), Positives = 89/149 (59%), Gaps = 15/149 (10%)
Query 4 PVFTPKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPIEFEKDD 63
P+F NL++A +E E K L K L S G + P+ FEKDD
Sbjct 766 PIFA--SNLELASASAEFGPEQQ----------KELNKALEVWS--VGPPLKPLMFEKDD 811
Query 64 DTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYKLV 123
D+NFH+DF+VAA++LR +NY IPP +R +SK I G+IIPAIATTTA V+GL+ +ELYK+V
Sbjct 812 DSNFHVDFVVAAASLRCQNYGIPPVNRAQSKRIVGQIIPAIATTTAAVAGLLGLELYKVV 871
Query 124 QGFTNTLEPYKNGFVNLALPFFGFSTPIA 152
G +++ +++LA + P A
Sbjct 872 SG-PRPRSAFRHSYLHLAENYLIRYMPFA 899
> SPBC16H5.03c
Length=628
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 34/78 (43%), Positives = 49/78 (62%), Gaps = 4/78 (5%)
Query 55 NPIEFEKDD-DTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSG 113
+ + F+KDD DT +DF+ AA+NLRA + I K +AG IIPAIATT A+++G
Sbjct 332 DDLSFDKDDKDT---LDFVAAAANLRAHVFGIQQLSEFDIKQMAGNIIPAIATTNAVIAG 388
Query 114 LVFIELYKLVQGFTNTLE 131
L + K++QG N L+
Sbjct 389 LCITQAIKVLQGDLNDLK 406
> At2g21470
Length=700
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 50/72 (69%), Gaps = 2/72 (2%)
Query 53 KINPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVS 112
+I + F+KDD ++F+ AA+N+RAE++ IP ++K IAG I+ A+ATT A+++
Sbjct 330 EIGHLTFDKDD--QLAVEFVTAAANIRAESFGIPLHSLFEAKGIAGNIVHAVATTNAIIA 387
Query 113 GLVFIELYKLVQ 124
GL+ IE K+++
Sbjct 388 GLIVIEAIKVLK 399
> CE25130
Length=456
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/66 (37%), Positives = 42/66 (63%), Gaps = 0/66 (0%)
Query 62 DDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYK 121
D D + F+ A +N+RA+ + IP + K +AG IIPAIA+T A+V+G++ E +
Sbjct 366 DKDHAIIMSFVAACANIRAKIFGIPMKSQFDIKAMAGNIIPAIASTNAIVAGIIVTEAVR 425
Query 122 LVQGFT 127
+++G T
Sbjct 426 VIEGST 431
> CE25131
Length=438
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/66 (37%), Positives = 42/66 (63%), Gaps = 0/66 (0%)
Query 62 DDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYK 121
D D + F+ A +N+RA+ + IP + K +AG IIPAIA+T A+V+G++ E +
Sbjct 348 DKDHAIIMSFVAACANIRAKIFGIPMKSQFDIKAMAGNIIPAIASTNAIVAGIIVTEAVR 407
Query 122 LVQGFT 127
+++G T
Sbjct 408 VIEGST 413
> Hs4885649
Length=640
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 52/95 (54%), Gaps = 0/95 (0%)
Query 31 DLNTHFKNLQKQLPSVSELAGVKINPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADR 90
D+ ++ + K + ++ K + E D D +DF+ +A+NLR + + R
Sbjct 314 DVKSYARLFSKSIETLRVHLAEKGDGAELIWDKDDPSAMDFVTSAANLRMHIFSMNMKSR 373
Query 91 HKSKLIAGRIIPAIATTTALVSGLVFIELYKLVQG 125
K +AG IIPAIATT A+++GL+ +E K++ G
Sbjct 374 FDIKSMAGNIIPAIATTNAVIAGLIVLEGLKILSG 408
> YDR390c
Length=636
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 30/60 (50%), Positives = 40/60 (66%), Gaps = 4/60 (6%)
Query 55 NPIEFEKDD-DTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSG 113
N IEF+KDD DT ++F+ A+N+R+ + IP K IAG IIPAIATT A+V+G
Sbjct 326 NHIEFDKDDADT---LEFVATAANIRSHIFNIPMKSVFDIKQIAGNIIPAIATTNAIVAG 382
> CE04374
Length=437
Score = 51.2 bits (121), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 41/74 (55%), Gaps = 0/74 (0%)
Query 62 DDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYK 121
D D H+++++ ++LRAE Y I DR + + RIIPA+A+T A+++ +E K
Sbjct 251 DADDPIHVEWVLERASLRAEKYNIRGVDRRLTSGVLKRIIPAVASTNAVIAASCALEALK 310
Query 122 LVQGFTNTLEPYKN 135
L ++ Y N
Sbjct 311 LATNIAKPIDNYLN 324
> 7295159_1
Length=731
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 45/71 (63%), Gaps = 2/71 (2%)
Query 55 NPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGL 114
+ + ++KDD +DF+ A +N+R+ ++I R + K +AG IIPAIATT A+ +G+
Sbjct 354 DTLAWDKDDQP--AMDFVAACANVRSHIFDIERKSRFEIKSMAGNIIPAIATTNAITAGI 411
Query 115 VFIELYKLVQG 125
+ +K+++
Sbjct 412 SVMRAFKVLEA 422
> Hs22043534
Length=168
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 17/32 (53%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 94 KLIAGRIIPAIATTTALVSGLVFIELYKLVQG 125
K +AG IIPAIATT A+++GL +E K++ G
Sbjct 8 KSVAGNIIPAIATTNAVIAGLTVLERLKILSG 39
> Hs19923744
Length=463
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 18/55 (32%), Positives = 31/55 (56%), Gaps = 0/55 (0%)
Query 68 HIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYKL 122
HI +I S RA Y I ++ + RIIPA+A+T A+++ + E++K+
Sbjct 277 HIQWIFQKSLERASQYNIRGVTYRLTQGVVKRIIPAVASTNAVIAAVCATEVFKI 331
> SPAC24H6.12c
Length=444
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 35/58 (60%), Gaps = 2/58 (3%)
Query 68 HIDFIVAASNLRAENYEIPPA--DRHKSKLIAGRIIPAIATTTALVSGLVFIELYKLV 123
HID++V S RA ++IP + +R + I RIIPA+A+T A+++ E K++
Sbjct 261 HIDWLVKRSIERANKFQIPSSSINRFFVQGIVKRIIPAVASTNAIIAASCCNEALKIL 318
> ECU02g1340
Length=429
Score = 33.5 bits (75), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 23/99 (23%), Positives = 51/99 (51%), Gaps = 7/99 (7%)
Query 22 NSENSNVPTDLN--THFKNLQKQLPSVSE-LAGVKINPIEFEKDDDTNFHIDFIVAASNL 78
N E+ + P+DL K+ +++ + E L + P F+KD+ +++I + +
Sbjct 201 NCEDMSTPSDLEKFRSCKDYKERTEKIVEILKNLDFGP--FDKDNRNT--LEYIYNVAYI 256
Query 79 RAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFI 117
R + P ++ IAG I+P+++T ++V+ L+ +
Sbjct 257 RGRCAGVEPTSFEEAITIAGNIVPSLSTINSIVASLMIL 295
> CE28275
Length=706
Score = 32.3 bits (72), Expect = 0.34, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 31/59 (52%), Gaps = 10/59 (16%)
Query 14 IAVNDSELNSENSNVPTDL---------NTHFKNLQKQLPSVSELAGVKINPIEFEKDD 63
+A +++ +N+ENS P+ N H++ QKQ S E ++ I+FE+DD
Sbjct 103 VAPSEASVNAENSRKPSKARLQSEMEIRNQHYQK-QKQYASTKETRAKRLTDIDFEEDD 160
> 7292913
Length=2050
Score = 32.3 bits (72), Expect = 0.34, Method: Composition-based stats.
Identities = 24/90 (26%), Positives = 42/90 (46%), Gaps = 6/90 (6%)
Query 57 IEFEKDDDTNFHI-DFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLV 115
IEFE DDDT F + S+L+ EI R KS +++ R+ P+ + ++ GL+
Sbjct 949 IEFEHDDDTVFFAHSYPYTYSDLQDYLMEIQRHPRKKSIVVSARVHPSETPASWMMKGLM 1008
Query 116 FIELYKLVQGFTNTLEPYKNGFVNLALPFF 145
+ G T + ++ F+ +P
Sbjct 1009 -----DFITGDTTVAKRLRHKFIFKLVPML 1033
> 7303261
Length=450
Score = 32.3 bits (72), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 38/83 (45%), Gaps = 0/83 (0%)
Query 53 KINPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVS 112
K NP D D HI +I + R+ + I + + IIPA+A+T A ++
Sbjct 240 KQNPFGVPLDGDDPQHIGWIYERALERSNEFNITGVTYRLVQGVVKHIIPAVASTNAAIA 299
Query 113 GLVFIELYKLVQGFTNTLEPYKN 135
+E++KL +++ Y N
Sbjct 300 AACALEVFKLATSCYDSMANYLN 322
> CE28274
Length=761
Score = 32.3 bits (72), Expect = 0.35, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 31/59 (52%), Gaps = 10/59 (16%)
Query 14 IAVNDSELNSENSNVPTDL---------NTHFKNLQKQLPSVSELAGVKINPIEFEKDD 63
+A +++ +N+ENS P+ N H++ QKQ S E ++ I+FE+DD
Sbjct 103 VAPSEASVNAENSRKPSKARLQSEMEIRNQHYQK-QKQYASTKETRAKRLTDIDFEEDD 160
> At5g19180
Length=477
Score = 30.8 bits (68), Expect = 0.98, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 100 IIPAIATTTALVSGLVFIELYKLVQGFTNTLEPY 133
IIPAIA+T A++S +E K+V + TL Y
Sbjct 307 IIPAIASTNAIISAACALETLKIVSACSKTLVNY 340
> CE25915
Length=824
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 30/64 (46%), Gaps = 0/64 (0%)
Query 2 EIPVFTPKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPIEFEK 61
E + P++N I + +LN V T + NL Q PS ++LA + F+K
Sbjct 439 EFDLLKPRENQSIKRAEQKLNYFGDIVDWCNETGYSNLSNQFPSATQLAKQHGDTYVFQK 498
Query 62 DDDT 65
D ++
Sbjct 499 DQNS 502
> At5g54730
Length=763
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 14/32 (43%), Positives = 21/32 (65%), Gaps = 1/32 (3%)
Query 99 RIIPAIATTTALVSGLVFIELYKLVQGFTNTL 130
RI+P I+T+ A V F L++L +GFTN +
Sbjct 376 RIMPTISTSRA-VKTTTFAHLFRLQRGFTNAV 406
> Hs20539754
Length=267
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 13/42 (30%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 53 KINPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSK 94
KI+P+ + + I+ + +AS L + + PP DR K K
Sbjct 157 KISPLGLSSEKNREAQIEVVTSASALIIKALKEPPRDRKKQK 198
> At1g10480
Length=211
Score = 28.5 bits (62), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 14/52 (26%), Positives = 30/52 (57%), Gaps = 0/52 (0%)
Query 11 NLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPIEFEKD 62
N ++ V + + +S +S + N ++L +Q + +L GV+ N I+F++D
Sbjct 135 NDQMGVYNEDWSSRSSQINFGNNDTCQDLNEQSGEMGKLYGVRPNMIQFQRD 186
> Hs20556540
Length=319
Score = 28.5 bits (62), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 19/45 (42%), Positives = 25/45 (55%), Gaps = 4/45 (8%)
Query 98 GRIIPAIATTTALVS---GLVFI-ELYKLVQGFTNTLEPYKNGFV 138
G I+ ++A TT V GL FI YK V G+ NT+E + N F
Sbjct 256 GIILISLAATTVTVVLFVGLGFIVRTYKPVHGYENTVESHVNSFC 300
> 7292326
Length=537
Score = 28.1 bits (61), Expect = 6.5, Method: Composition-based stats.
Identities = 11/37 (29%), Positives = 25/37 (67%), Gaps = 0/37 (0%)
Query 8 PKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLP 44
P Q+++I ++ S+ NS++ ++ L+ F NL++ +P
Sbjct 345 PTQSIQINLSSSKENSQSDSISETLSESFLNLEENIP 381
> CE05426
Length=686
Score = 28.1 bits (61), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 28/96 (29%), Positives = 40/96 (41%), Gaps = 15/96 (15%)
Query 60 EKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTA--LVSGLVFI 117
EKDDDT + F + N+ K KL + R +P + T ++ L +
Sbjct 274 EKDDDTIY--SFTIDIKESEVSNF--------KEKLESERFLPTLYDTNYDNYLNELKQV 323
Query 118 EL---YKLVQGFTNTLEPYKNGFVNLALPFFGFSTP 150
L +K Q F NT + YK L + F STP
Sbjct 324 LLGFNWKQHQHFLNTFKQYKTEIEGLKIHFLRVSTP 359
> Hs22060509
Length=1093
Score = 28.1 bits (61), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 1 EEIPVFTPKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPS 45
E++P+ P Q L N E++S ++N P + N + L + PS
Sbjct 255 EDLPLRHPAQALGKPKNQQEVSSASNNTPEEQNDFMQQLPSRCPS 299
> Hs14769393
Length=1093
Score = 28.1 bits (61), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 1 EEIPVFTPKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPS 45
E++P+ P Q L N E++S ++N P + N + L + PS
Sbjct 255 EDLPLRHPAQALGKPKNQQEVSSASNNTPEEQNDFMQQLPSRCPS 299
> Hs18587236
Length=466
Score = 27.7 bits (60), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 27/121 (22%), Positives = 50/121 (41%), Gaps = 29/121 (23%)
Query 1 EEIPVFTPKQN-----LKIAVNDSE-------------LNSENSNVP-----TDLNTHF- 36
EE+ F P+ + ++A+ DSE N P DL+ H
Sbjct 262 EELAAFCPQLDDSTVARELAITDSEHSDAEVSCTDNGTFNLSRGQTPLTEGSEDLDGHSD 321
Query 37 --KNLQKQLPSVSELAGVKINPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSK 94
++ + LP + + ++P + +DDT+ + ++ S AE + PPA R ++
Sbjct 322 PEESFARDLP---DFPSINMDPAGLDDEDDTSIGMPSLMYRSPPGAEEPQAPPASRDEAA 378
Query 95 L 95
L
Sbjct 379 L 379
Lambda K H
0.317 0.135 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2070320142
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40