bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1814_orf1
Length=179
Score E
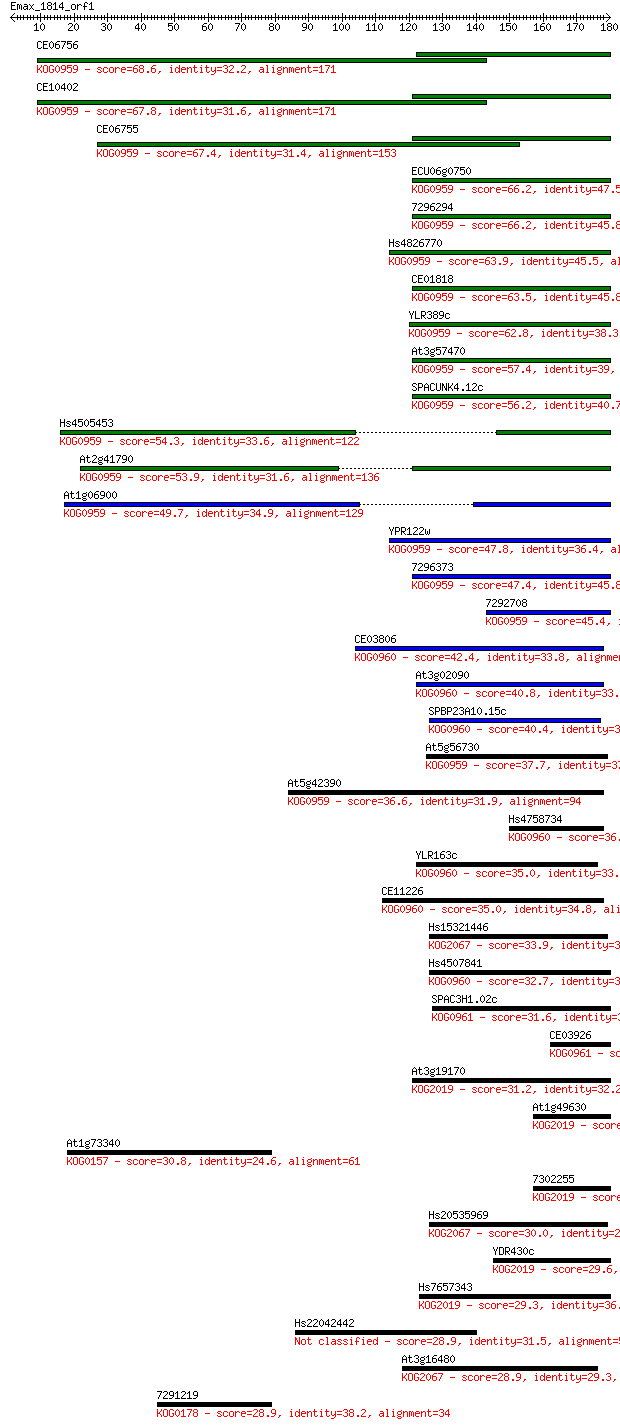
Sequences producing significant alignments: (Bits) Value
CE06756 68.6 7e-12
CE10402 67.8 1e-11
CE06755 67.4 1e-11
ECU06g0750 66.2 3e-11
7296294 66.2 3e-11
Hs4826770 63.9 2e-10
CE01818 63.5 2e-10
YLR389c 62.8 4e-10
At3g57470 57.4 2e-08
SPACUNK4.12c 56.2 3e-08
Hs4505453 54.3 1e-07
At2g41790 53.9 2e-07
At1g06900 49.7 3e-06
YPR122w 47.8 1e-05
7296373 47.4 2e-05
7292708 45.4 6e-05
CE03806 42.4 5e-04
At3g02090 40.8 0.001
SPBP23A10.15c 40.4 0.002
At5g56730 37.7 0.014
At5g42390 36.6 0.025
Hs4758734 36.2 0.036
YLR163c 35.0 0.070
CE11226 35.0 0.078
Hs15321446 33.9 0.16
Hs4507841 32.7 0.41
SPAC3H1.02c 31.6 0.82
CE03926 31.6 0.98
At3g19170 31.2 1.3
At1g49630 30.8 1.3
At1g73340 30.8 1.5
7302255 30.4 2.1
Hs20535969 30.0 2.3
YDR430c 29.6 3.0
Hs7657343 29.3 4.3
Hs22042442 28.9 5.5
At3g16480 28.9 5.6
7291219 28.9 6.3
> CE06756
Length=845
Score = 68.6 bits (166), Expect = 7e-12, Method: Composition-based stats.
Identities = 29/58 (50%), Positives = 41/58 (70%), Gaps = 0/58 (0%)
Query 122 RHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
R +L+NG+ + V P ++++ ++A G L DP ++PGLAHF EHMLFLGTSKYP
Sbjct 27 RGLELTNGLRVLLVSDPTTDKSAVSLAVKAGHLMDPWELPGLAHFCEHMLFLGTSKYP 84
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 32/135 (23%), Positives = 59/135 (43%), Gaps = 17/135 (12%)
Query 9 MAQQSQIDFHTTQPSSSIMDEAARLAHNLLTYEPY-HVVAGDSLLIDADPRLTNQLLQEM 67
+A+ S I+F + + A ++A NL Y P+ H+++ LL +P +LL +
Sbjct 377 LAELSAIEFRF-KDREPLTKNAIKVARNL-QYIPFEHILSSRYLLTKYNPERIKELLSTL 434
Query 68 SPSKAIIAFSDPDFTSKVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRHYQLS 127
+PS ++ F + + +P YG + +V D+ SP + Y+ +
Sbjct 435 TPSNMLVRVVSKKFKEQ-EGNTNEPVYGTEMKVTDI-------------SPEKMKKYENA 480
Query 128 NGMHAIAVHHPRSNE 142
A+H P NE
Sbjct 481 LKTSHHALHLPEKNE 495
> CE10402
Length=1067
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 28/59 (47%), Positives = 41/59 (69%), Gaps = 0/59 (0%)
Query 121 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+R +L+NG+ + V P ++++ A+ G L DP ++PGLAHF EHMLFLGT+KYP
Sbjct 85 YRGLELTNGIRVLLVSDPTTDKSAAALDVKVGHLMDPWELPGLAHFCEHMLFLGTAKYP 143
Score = 34.7 bits (78), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 32/138 (23%), Positives = 57/138 (41%), Gaps = 23/138 (16%)
Query 9 MAQQSQIDFH---TTQPSSSIMDEAARLAHNLLTYEPY-HVVAGDSLLIDADPRLTNQLL 64
+A+ S + F QP + ++ AA L Y P+ H+++ LL +P +LL
Sbjct 436 LAELSAVKFRFKDKEQPMTMAINVAASLQ-----YIPFEHILSSRYLLTKYEPERIKELL 490
Query 65 QEMSPSKAIIAFSDPDFTSKVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRHY 124
+SP+ + F + + +P YG + +V D+ SP + Y
Sbjct 491 SMLSPANMQVRVVSQKFKGQ-EGNTNEPVYGTEMKVTDI-------------SPETMKKY 536
Query 125 QLSNGMHAIAVHHPRSNE 142
+ + A+H P NE
Sbjct 537 ENALKTSHHALHLPEKNE 554
> CE06755
Length=980
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 28/59 (47%), Positives = 41/59 (69%), Gaps = 0/59 (0%)
Query 121 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+R +L+NG+ + V P ++++ A+ G L DP ++PGLAHF EHMLFLGT+KYP
Sbjct 26 YRGLELTNGIRVLLVSDPTTDKSAAALDVKVGHLMDPWELPGLAHFCEHMLFLGTAKYP 84
Score = 37.4 bits (85), Expect = 0.015, Method: Composition-based stats.
Identities = 29/126 (23%), Positives = 51/126 (40%), Gaps = 17/126 (13%)
Query 27 MDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKVD 86
M A +A +L H+++ LL +P +LL ++PS ++ F + +
Sbjct 367 MKMAINIAASLQYIPIEHILSSRYLLTKYEPERIKELLSTLTPSNMLVRVVSQKFKEQ-E 425
Query 87 SFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRHYQLSNGMHAIAVHHPRSNEAGFA 146
+P YG + +V D+ SP + Y+ + A+H P NE
Sbjct 426 GNTNEPVYGTEMKVTDI-------------SPEKMKKYENALKTSHHALHLPEKNE---Y 469
Query 147 VAANTG 152
+A N G
Sbjct 470 IATNFG 475
> ECU06g0750
Length=882
Score = 66.2 bits (160), Expect = 3e-11, Method: Composition-based stats.
Identities = 28/59 (47%), Positives = 38/59 (64%), Gaps = 0/59 (0%)
Query 121 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+ + ++ NGM A+ + P ++ AV+ GS DP D GLAHFLEHMLF+GT KYP
Sbjct 27 YEYVEIPNGMRALIMSDPGLDKCSCAVSVRVGSFDDPADAQGLAHFLEHMLFMGTEKYP 85
> 7296294
Length=990
Score = 66.2 bits (160), Expect = 3e-11, Method: Composition-based stats.
Identities = 27/59 (45%), Positives = 39/59 (66%), Gaps = 0/59 (0%)
Query 121 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+R QL NG+ + + P ++ + A++ G + DP ++PGLAHF EHMLFLGT KYP
Sbjct 37 YRGLQLENGLKVLLISDPNTDVSAAALSVQVGHMSDPTNLPGLAHFCEHMLFLGTEKYP 95
> Hs4826770
Length=1019
Score = 63.9 bits (154), Expect = 2e-10, Method: Composition-based stats.
Identities = 30/69 (43%), Positives = 45/69 (65%), Gaps = 3/69 (4%)
Query 114 LTASPNAFRHY---QLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHM 170
+T SP R Y +L+NG+ + + P ++++ A+ + GSL DP ++ GL+HF EHM
Sbjct 54 ITKSPEDKREYRGLELANGIKVLLMSDPTTDKSSAALDVHIGSLSDPPNIAGLSHFCEHM 113
Query 171 LFLGTSKYP 179
LFLGT KYP
Sbjct 114 LFLGTKKYP 122
> CE01818
Length=745
Score = 63.5 bits (153), Expect = 2e-10, Method: Composition-based stats.
Identities = 27/59 (45%), Positives = 39/59 (66%), Gaps = 0/59 (0%)
Query 121 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+R +L+NG+ + V ++ + A+ G L DP ++PGLAHF EHMLFLGT+KYP
Sbjct 27 YRGLELTNGLRVLLVSDSKTRVSAVALDVKVGHLMDPWELPGLAHFCEHMLFLGTAKYP 85
> YLR389c
Length=988
Score = 62.8 bits (151), Expect = 4e-10, Method: Composition-based stats.
Identities = 23/60 (38%), Positives = 43/60 (71%), Gaps = 0/60 (0%)
Query 120 AFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
++R +L N + A+ + P++++A ++ N G+ DP+++PGLAHF EH+LF+G+ K+P
Sbjct 73 SYRFIELPNKLKALLIQDPKADKAAASLDVNIGAFEDPKNLPGLAHFCEHLLFMGSEKFP 132
> At3g57470
Length=989
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 23/59 (38%), Positives = 35/59 (59%), Gaps = 0/59 (0%)
Query 121 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+R L N + + + P +++ ++ + GS DPE + GLAHFLEHMLF + KYP
Sbjct 27 YRRIVLKNSLEVLLISDPETDKCAASMNVSVGSFTDPEGLEGLAHFLEHMLFYASEKYP 85
> SPACUNK4.12c
Length=969
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 24/59 (40%), Positives = 37/59 (62%), Gaps = 0/59 (0%)
Query 121 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+R +L N + + V P ++ A A+ + GS +P ++ GLAHF EH+LF+GT KYP
Sbjct 24 YRLIKLENDLEVLLVRDPETDNASAAIDVHIGSQSNPRELLGLAHFCEHLLFMGTKKYP 82
> Hs4505453
Length=1219
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 21/34 (61%), Positives = 27/34 (79%), Gaps = 0/34 (0%)
Query 146 AVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
A+ GS DP+D+PGLAHFLEHM+F+G+ KYP
Sbjct 282 ALCVGVGSFADPDDLPGLAHFLEHMVFMGSLKYP 315
Score = 32.7 bits (73), Expect = 0.44, Method: Composition-based stats.
Identities = 20/88 (22%), Positives = 39/88 (44%), Gaps = 3/88 (3%)
Query 16 DFHTTQPSSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIA 75
+FH Q + ++ + N+ Y ++ GD LL + P + + L ++ P KA +
Sbjct 620 EFHY-QEQTDPVEYVENMCENMQLYPLQDILTGDQLLFEYKPEVIGEALNQLVPQKANLV 678
Query 76 FSDPDFTSKVDSFETDPYYGVQFRVLDL 103
K D E ++G Q+ + D+
Sbjct 679 LLSGANEGKCDLKEK--WFGTQYSIEDI 704
> At2g41790
Length=970
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 36/59 (61%), Gaps = 0/59 (0%)
Query 121 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+R L N + + + P +++ +++ + GS DP+ + GLAHFLEHMLF + KYP
Sbjct 25 YRMIVLKNLLQVLLISDPDTDKCAASMSVSVGSFSDPQGLEGLAHFLEHMLFYASEKYP 83
Score = 32.3 bits (72), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 35/77 (45%), Gaps = 6/77 (7%)
Query 22 PSSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDF 81
P S I+D +A N+ Y + G SL +P + +++ E+SPS I + F
Sbjct 391 PMSYIVD----IASNMQIYPTKDWLVGSSLPTKFNPAIVQKVVDELSPSNFRIFWESQKF 446
Query 82 TSKVDSFETDPYYGVQF 98
+ D E P+Y +
Sbjct 447 EGQTDKAE--PWYNTAY 461
> At1g06900
Length=1023
Score = 49.7 bits (117), Expect = 3e-06, Method: Composition-based stats.
Identities = 19/41 (46%), Positives = 30/41 (73%), Gaps = 0/41 (0%)
Query 139 RSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
++ +A A+ + GS DP + GLAHFLEHMLF+G++++P
Sbjct 103 QTKKAAAAMCVSMGSFLDPPEAQGLAHFLEHMLFMGSTEFP 143
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 26/88 (29%), Positives = 41/88 (46%), Gaps = 5/88 (5%)
Query 17 FHTTQPSSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAF 76
F QP+ D AA L+ N+L Y HV+ GD + DP+L L+ +P I
Sbjct 448 FAEEQPAD---DYAAELSENMLAYPVEHVIYGDYVYQTWDPKLIEDLMGFFTPQNMRIDV 504
Query 77 SDPDFTSKVDSFETDPYYGVQFRVLDLP 104
S + F+ +P++G + D+P
Sbjct 505 VSKSIKS--EEFQQEPWFGSSYIEEDVP 530
> YPR122w
Length=1208
Score = 47.8 bits (112), Expect = 1e-05, Method: Composition-based stats.
Identities = 24/67 (35%), Positives = 37/67 (55%), Gaps = 1/67 (1%)
Query 114 LTASPNAFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLF- 172
L+ S + +L NG+ A+ + P + ++ TGS DP+D+ GLAH EHM+
Sbjct 17 LSYSNRTHKVCKLPNGILALIISDPTDTSSSCSLTVCTGSHNDPKDIAGLAHLCEHMILS 76
Query 173 LGTSKYP 179
G+ KYP
Sbjct 77 AGSKKYP 83
> 7296373
Length=908
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 38/81 (46%), Gaps = 22/81 (27%)
Query 121 FRHYQLSNGMHAIAVH---------HPRSNE-------------AGFAVAANTGSLYDPE 158
+R LSNG+ A+ + H S E A AV GS +P+
Sbjct 50 YRALTLSNGLRAMLISDSYIDEPSIHRASRESLNSSTENFNGKLAACAVLVGVGSFSEPQ 109
Query 159 DVPGLAHFLEHMLFLGTSKYP 179
GLAHF+EHM+F+G+ K+P
Sbjct 110 QYQGLAHFVEHMIFMGSEKFP 130
> 7292708
Length=1077
Score = 45.4 bits (106), Expect = 6e-05, Method: Composition-based stats.
Identities = 19/37 (51%), Positives = 25/37 (67%), Gaps = 0/37 (0%)
Query 143 AGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
A A+ + GS +P GLAHFLEHM+F+G+ KYP
Sbjct 94 AACALLIDYGSFAEPTKYQGLAHFLEHMIFMGSEKYP 130
> CE03806
Length=485
Score = 42.4 bits (98), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 25/74 (33%), Positives = 34/74 (45%), Gaps = 10/74 (13%)
Query 104 PQHHAVAMAVLTASPNAFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGL 163
P+ V ++T PN FR +A + + A V + GS Y+ E G
Sbjct 49 PKSVFVPETIVTTLPNGFR----------VATENTGGSTATIGVFIDAGSRYENEKNNGT 98
Query 164 AHFLEHMLFLGTSK 177
AHFLEHM F GT +
Sbjct 99 AHFLEHMAFKGTPR 112
> At3g02090
Length=531
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 29/56 (51%), Gaps = 0/56 (0%)
Query 122 RHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSK 177
R L NG+ + + A V + GS ++ ++ G AHFLEHM+F GT +
Sbjct 98 RVTTLPNGLRVATESNLSAKTATVGVWIDAGSRFESDETNGTAHFLEHMIFKGTDR 153
> SPBP23A10.15c
Length=457
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 25/51 (49%), Gaps = 0/51 (0%)
Query 126 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTS 176
L NG+ HHP + A V + GS + G AHFLEH+ F GT
Sbjct 27 LKNGLTVATEHHPYAQTATVLVGVDAGSRAETAKNNGAAHFLEHLAFKGTK 77
> At5g56730
Length=956
Score = 37.7 bits (86), Expect = 0.014, Method: Composition-based stats.
Identities = 20/59 (33%), Positives = 33/59 (55%), Gaps = 9/59 (15%)
Query 125 QLSNGMHAIAVHHPRSN-----EAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 178
+L NG+ +++ R N A A+A GS+ + ED G+AH +EH+ F T++Y
Sbjct 44 RLDNGL----IYYVRRNSKPRMRAALALAVKVGSVLEEEDQRGVAHIVEHLAFSATTRY 98
> At5g42390
Length=1265
Score = 36.6 bits (83), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 46/97 (47%), Gaps = 22/97 (22%)
Query 84 KVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRHYQLSNGMHAIAVHH---PRS 140
++DS E + + G + LP H P R QL NG+ + + + P
Sbjct 175 EIDSAELEAFLGCE-----LPSH-----------PKLHRG-QLKNGLRYLILPNKVPPNR 217
Query 141 NEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSK 177
EA V + GS+ + ED G+AH +EH+ FLG+ K
Sbjct 218 FEAHMEV--HVGSIDEEEDEQGIAHMIEHVAFLGSKK 252
> Hs4758734
Length=489
Score = 36.2 bits (82), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 16/28 (57%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 150 NTGSLYDPEDVPGLAHFLEHMLFLGTSK 177
+ GS Y+ E G AHFLEHM F GT K
Sbjct 86 DAGSRYENEKNNGTAHFLEHMAFKGTKK 113
> YLR163c
Length=462
Score = 35.0 bits (79), Expect = 0.070, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 27/54 (50%), Gaps = 0/54 (0%)
Query 122 RHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGT 175
R +L NG+ + P ++ A + + GS + G AHFLEH+ F GT
Sbjct 27 RTSKLPNGLTIATEYIPNTSSATVGIFVDAGSRAENVKNNGTAHFLEHLAFKGT 80
> CE11226
Length=471
Score = 35.0 bits (79), Expect = 0.078, Method: Compositional matrix adjust.
Identities = 23/66 (34%), Positives = 29/66 (43%), Gaps = 10/66 (15%)
Query 112 AVLTASPNAFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHML 171
A +T N FR NG S A V TGS ++ E G+AHFLE ++
Sbjct 38 AEVTTLKNGFRVVTEDNG----------SATATVGVWIETGSRFENEKNNGVAHFLERLI 87
Query 172 FLGTSK 177
GT K
Sbjct 88 HKGTGK 93
> Hs15321446
Length=525
Score = 33.9 bits (76), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 29/53 (54%), Gaps = 1/53 (1%)
Query 126 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 178
L NG+ +A + + N+GS Y+ + + G+AHFLE + F T+++
Sbjct 72 LDNGLR-VASQNKFGQFCTVGILINSGSRYEAKYLSGIAHFLEKLAFSSTARF 123
> Hs4507841
Length=480
Score = 32.7 bits (73), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 24/54 (44%), Gaps = 1/54 (1%)
Query 126 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
L NG+ +A V + GS ++ E G +FLEH+ F GT P
Sbjct 53 LDNGLR-VASEQSSQPTCTVGVWIDVGSRFETEKNNGAGYFLEHLAFKGTKNRP 105
> SPAC3H1.02c
Length=1036
Score = 31.6 bits (70), Expect = 0.82, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 24/53 (45%), Gaps = 5/53 (9%)
Query 127 SNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+ G +V P S G V A + D G H LEH+ F+G+ KYP
Sbjct 28 ATGFSVASVKTPTSRLQGSFVVAT-----EAHDNLGCPHTLEHLCFMGSKKYP 75
> CE03926
Length=995
Score = 31.6 bits (70), Expect = 0.98, Method: Compositional matrix adjust.
Identities = 11/18 (61%), Positives = 15/18 (83%), Gaps = 0/18 (0%)
Query 162 GLAHFLEHMLFLGTSKYP 179
GL H LEH++F+G+ KYP
Sbjct 57 GLPHTLEHLVFMGSKKYP 74
> At3g19170
Length=1052
Score = 31.2 bits (69), Expect = 1.3, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 29/59 (49%), Gaps = 7/59 (11%)
Query 121 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
F+H + G ++V + N+ F V T P+D G+ H LEH + G+ KYP
Sbjct 97 FKHKK--TGCEVMSVSNEDENKV-FGVVFRT----PPKDSTGIPHILEHSVLCGSRKYP 148
> At1g49630
Length=1076
Score = 30.8 bits (68), Expect = 1.3, Method: Composition-based stats.
Identities = 11/23 (47%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 157 PEDVPGLAHFLEHMLFLGTSKYP 179
P+D G+ H LEH + G+ KYP
Sbjct 153 PKDSTGIPHILEHSVLCGSRKYP 175
> At1g73340
Length=512
Score = 30.8 bits (68), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 15/61 (24%), Positives = 30/61 (49%), Gaps = 0/61 (0%)
Query 18 HTTQPSSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFS 77
H + + +++E RLA +LT++ Y + +ID RL + M +K +++
Sbjct 334 HCPKAMTQLLEEHDRLAGGMLTWQDYKTMDFTQCVIDETLRLGGIAIWLMREAKEDVSYQ 393
Query 78 D 78
D
Sbjct 394 D 394
> 7302255
Length=1112
Score = 30.4 bits (67), Expect = 2.1, Method: Composition-based stats.
Identities = 12/23 (52%), Positives = 14/23 (60%), Gaps = 0/23 (0%)
Query 157 PEDVPGLAHFLEHMLFLGTSKYP 179
P D GL H LEH+ G+ KYP
Sbjct 120 PFDSTGLPHILEHLSLCGSQKYP 142
> Hs20535969
Length=417
Score = 30.0 bits (66), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 26/53 (49%), Gaps = 1/53 (1%)
Query 126 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 178
L NG +A + + N+GS Y+ + G+AHFLE + F +++
Sbjct 20 LDNGFR-VASQNKFGQFCIVGILINSGSRYEATYLSGIAHFLEKLAFSSNARF 71
> YDR430c
Length=989
Score = 29.6 bits (65), Expect = 3.0, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 19/35 (54%), Gaps = 4/35 (11%)
Query 145 FAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
F++A T +P D G+ H LEH G+ KYP
Sbjct 68 FSIAFKT----NPPDSTGVPHILEHTTLCGSVKYP 98
> Hs7657343
Length=1038
Score = 29.3 bits (64), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 21/80 (26%), Positives = 30/80 (37%), Gaps = 23/80 (28%)
Query 123 HYQLSNGMHAIAVHHPRSNEAGFAVAA-----NTGSLY------------------DPED 159
Y+L + +H V+ S F A +TG+ Y P D
Sbjct 39 QYKLGDKIHGFTVNQVTSVPELFLTAVKLTHDDTGARYLHLAREDTNNLFSVQFRTTPMD 98
Query 160 VPGLAHFLEHMLFLGTSKYP 179
G+ H LEH + G+ KYP
Sbjct 99 STGVPHILEHTVLCGSQKYP 118
> Hs22042442
Length=746
Score = 28.9 bits (63), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 26/55 (47%), Gaps = 7/55 (12%)
Query 86 DSFETDPYYGVQFRVLDLPQHH-AVAMAVLTASPNAFRHYQLSNGMHAIAVHHPR 139
DS E+ PYY + + P HH V M + AF +Y SN + ++ P+
Sbjct 533 DSPESQPYYSSIWSAFNNPTHHRTVTMTI------AFANYGWSNSSASTSLREPQ 581
> At3g16480
Length=448
Score = 28.9 bits (63), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 25/58 (43%), Gaps = 1/58 (1%)
Query 118 PNAFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGT 175
P+ + L NG+ IA + A + + GS+Y+ G H LE M F T
Sbjct 20 PSKLKTTTLPNGL-TIATEMSPNPAASIGLYVDCGSIYETPQFRGATHLLERMAFKST 76
> 7291219
Length=264
Score = 28.9 bits (63), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 45 VVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSD 78
++A D +L+ A+ R TN+LL PS+ I +D
Sbjct 37 ILAEDGILLAAECRSTNKLLDSAIPSEKIYRLND 70
Lambda K H
0.318 0.132 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2815454148
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40