bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1811_orf1
Length=141
Score E
Sequences producing significant alignments: (Bits) Value
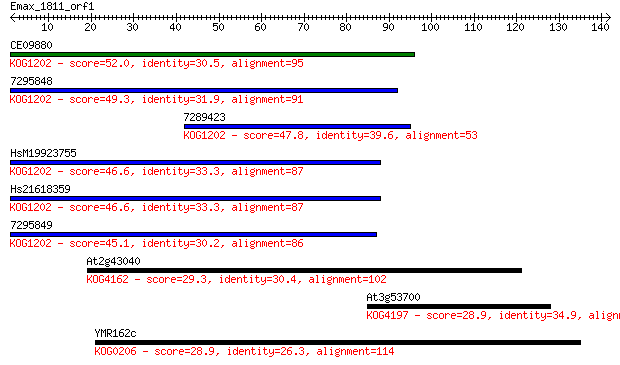
CE09880 52.0 4e-07
7295848 49.3 2e-06
7289423 47.8 8e-06
HsM19923755 46.6 2e-05
Hs21618359 46.6 2e-05
7295849 45.1 5e-05
At2g43040 29.3 2.2
At3g53700 28.9 3.7
YMR162c 28.9 3.7
> CE09880
Length=2586
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 29/98 (29%), Positives = 54/98 (55%), Gaps = 3/98 (3%)
Query 1 FLQVPSPVLSSLIKSNLEDASRKSDDDVLEQLCAHLCVDKRSR---DETIGEAGLDSMTV 57
FL P++SS IK+ L D++ + L V+ S+ D + + GLDS+
Sbjct 2165 FLSWNHPIVSSFIKAELGSKKNVGGGDLMATIAHILGVNDISQLNADANLSDLGLDSLMG 2224
Query 58 VEIQQRLERDYEVMFTVADVKRITVGELKDSRDGKREG 95
VEI+Q LERD++++ ++ +++ +T+ +L+ D G
Sbjct 2225 VEIKQALERDHDIVLSMKEIRTLTLNKLQQLADQGGTG 2262
> 7295848
Length=2438
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/98 (29%), Positives = 52/98 (53%), Gaps = 11/98 (11%)
Query 1 FLQVPSPVLSSLIKSNLEDASRKSDDDVLEQLCAHLCVDKRSRD-------ETIGEAGLD 53
FLQ P PV++S++ + RKSD L A + RD ++ + G+D
Sbjct 2025 FLQQPHPVVASMVVAE----KRKSDQSAGVSLIATIANILGLRDTKNIQDGASLADLGMD 2080
Query 54 SMTVVEIQQRLERDYEVMFTVADVKRITVGELKDSRDG 91
S+ EI+Q LER+++++ + +++++T G LK G
Sbjct 2081 SLMSAEIKQTLERNFDIVLSAQEIRQLTFGALKAMDGG 2118
> 7289423
Length=2176
Score = 47.8 bits (112), Expect = 8e-06, Method: Composition-based stats.
Identities = 21/53 (39%), Positives = 35/53 (66%), Gaps = 0/53 (0%)
Query 42 SRDETIGEAGLDSMTVVEIQQRLERDYEVMFTVADVKRITVGELKDSRDGKRE 94
S T+ E G+DS+ VEI+Q LERD+E++ T D++ +T +L++ D K +
Sbjct 1907 SLSTTLSEMGMDSLMAVEIKQTLERDFELILTPQDLRHLTFQKLQEFIDAKEK 1959
> HsM19923755
Length=2509
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 29/93 (31%), Positives = 55/93 (59%), Gaps = 7/93 (7%)
Query 1 FLQVPSPVLSSLIKSNLEDASRKSDD--DVLEQLCAHLC----VDKRSRDETIGEAGLDS 54
FL P VLSS + + A R D D++E + AH+ + + D ++ + GLDS
Sbjct 2096 FLNQPHMVLSSFVLAEKAAAYRDRDSQRDLVEAV-AHILGIRDLAAVNLDSSLADLGLDS 2154
Query 55 MTVVEIQQRLERDYEVMFTVADVKRITVGELKD 87
+ VE++Q LER+ ++ +V +V+++T+ +L++
Sbjct 2155 LMSVEVRQTLERELNLVLSVREVRQLTLRKLQE 2187
> Hs21618359
Length=2509
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 29/93 (31%), Positives = 55/93 (59%), Gaps = 7/93 (7%)
Query 1 FLQVPSPVLSSLIKSNLEDASRKSDD--DVLEQLCAHLC----VDKRSRDETIGEAGLDS 54
FL P VLSS + + A R D D++E + AH+ + + D ++ + GLDS
Sbjct 2096 FLNQPHMVLSSFVLAEKAAAYRDRDSQRDLVEAV-AHILGIRDLAAVNLDSSLADLGLDS 2154
Query 55 MTVVEIQQRLERDYEVMFTVADVKRITVGELKD 87
+ VE++Q LER+ ++ +V +V+++T+ +L++
Sbjct 2155 LMSVEVRQTLERELNLVLSVREVRQLTLRKLQE 2187
> 7295849
Length=2409
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 49/89 (55%), Gaps = 3/89 (3%)
Query 1 FLQVPSPVLSSLI---KSNLEDASRKSDDDVLEQLCAHLCVDKRSRDETIGEAGLDSMTV 57
FLQ P PV++S++ K E A+R S + + V S T+ + G+DS+
Sbjct 2004 FLQQPHPVMASMVVAEKRKTEQANRLSLIATIANIMGLRDVKSVSDKTTLFDLGMDSLMS 2063
Query 58 VEIQQRLERDYEVMFTVADVKRITVGELK 86
EI+Q LER ++++ + +++++T L+
Sbjct 2064 TEIKQTLERHFDLVLSAQEIRQLTFSALR 2092
> At2g43040
Length=666
Score = 29.3 bits (64), Expect = 2.2, Method: Composition-based stats.
Identities = 31/105 (29%), Positives = 50/105 (47%), Gaps = 15/105 (14%)
Query 19 DASRKSDDDVLEQLCAH-LCVDKRSRDETIGEAGLDSMTVVEIQQRLERDYEVMFTVADV 77
D S K +D+++ QLCA+ +C+ T EA LD + E + L + F A
Sbjct 8 DFSEKGEDEIVRQLCANGICMKT-----TEVEAKLDEGNIQEAESSLREGLSLNFEEA-- 60
Query 78 KRITVGELKDSRDGKREGLKQISE--DLKRARMHLSTIKFEIPTE 120
R +G L+ R G EG ++ E DL+ A + ++ +P E
Sbjct 61 -RALLGRLEYQR-GNLEGALRVFEGIDLQAA---IQRLQVSVPLE 100
> At3g53700
Length=754
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 85 LKDSRDGKREGLKQISEDLKRARMHLSTIKFEIPTEPYTVVNL 127
L+ R G + +K+I ED+K +R + T F I E Y L
Sbjct 91 LRLGRSGSFDDMKKILEDMKSSRCEMGTSTFLILIESYAQFEL 133
> YMR162c
Length=1656
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 30/122 (24%), Positives = 54/122 (44%), Gaps = 12/122 (9%)
Query 21 SRKSDDDVLEQLCAHLCVDKRSRDETIGEAGLDSMTVVEIQQRL---ERDYEVMFTVADV 77
S+K+ L H C+ K++ +E+IGE ++ + + L RD + +
Sbjct 725 SQKAKFFFLSLALCHSCLPKKTHNESIGEDSIEYQSSSPDELALVTAARDLGYIVLNRNA 784
Query 78 KRITVGELKDSRDGK-----REGLKQISEDLKRARMHLSTIKFEIPTEPYTVVNLISKTN 132
+ +T+ D DG+ E L I + +R RM ++ +P +P V+ LI K
Sbjct 785 QILTIKTFPDGFDGEAKLENYEILNYIDFNSQRKRM---SVLVRMPNQPNQVL-LICKGA 840
Query 133 SN 134
N
Sbjct 841 DN 842
Lambda K H
0.315 0.132 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1608078824
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40