bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1807_orf1
Length=143
Score E
Sequences producing significant alignments: (Bits) Value
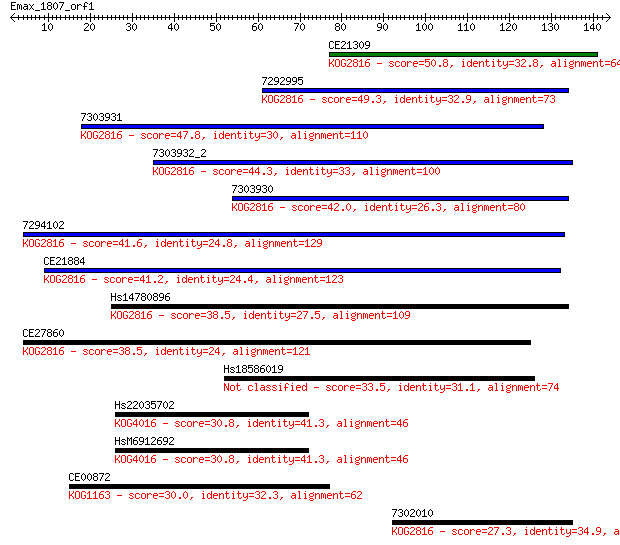
CE21309 50.8 8e-07
7292995 49.3 2e-06
7303931 47.8 7e-06
7303932_2 44.3 7e-05
7303930 42.0 4e-04
7294102 41.6 5e-04
CE21884 41.2 6e-04
Hs14780896 38.5 0.004
CE27860 38.5 0.004
Hs18586019 33.5 0.14
Hs22035702 30.8 0.86
HsM6912692 30.8 0.87
CE00872 30.0 1.4
7302010 27.3 8.8
> CE21309
Length=452
Score = 50.8 bits (120), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 47/64 (73%), Gaps = 0/64 (0%)
Query 77 GVRSRLSKIINEDELGKLFALLAIVEAVTPCVASLFYSILFAVSIDIYPGLVFMLAASSL 136
G R+ +S +I+ E GK+F++++++E +T VA+ Y+ ++ ++ +PGL+++++A +L
Sbjct 349 GFRAFISSLIDMQEQGKIFSVISLLEGITTLVATSIYNNVYPNTLTFFPGLLYLISAVTL 408
Query 137 LVPL 140
L+PL
Sbjct 409 LIPL 412
> 7292995
Length=599
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 46/73 (63%), Gaps = 0/73 (0%)
Query 61 LYLSFILGSISPFSFIGVRSRLSKIINEDELGKLFALLAIVEAVTPCVASLFYSILFAVS 120
+YL ++ + +FI +RS +K++++DELGK+ +L + EA+ P V + Y+ L+A +
Sbjct 421 MYLGGLVEIFNGTAFIAMRSIATKLVSKDELGKVNSLFGVAEALMPMVFAPMYTTLYAAT 480
Query 121 IDIYPGLVFMLAA 133
+ + PG F+L
Sbjct 481 LRVLPGAFFLLGG 493
> 7303931
Length=533
Score = 47.8 bits (112), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 59/110 (53%), Gaps = 2/110 (1%)
Query 18 LMGLLLPIFQNVFKFGDMRLALMGVLSLMAMCILRGAWQHVPGLYLSFILGSISPFSFIG 77
L+G L I + VF+ + LAL+ S + + +G +YLS LG S
Sbjct 391 LIGFL--ILRKVFRLSVVTLALLAFFSEILNNLAKGFATMPWHMYLSVTLGVFRSISGPM 448
Query 78 VRSRLSKIINEDELGKLFALLAIVEAVTPCVASLFYSILFAVSIDIYPGL 127
R+ +S I+ +LGK+F++ ++++ P VA+ Y++++ S+ YPGL
Sbjct 449 CRTIVSNIVPPSDLGKIFSIKNVLQSFAPFVAAPLYTLIYKRSLTTYPGL 498
> 7303932_2
Length=493
Score = 44.3 bits (103), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 33/107 (30%), Positives = 54/107 (50%), Gaps = 11/107 (10%)
Query 35 MRLALMGVLSLMAMCILRGA-------WQHVPGLYLSFILGSISPFSFIGVRSRLSKIIN 87
M + M +LSL A C+L WQ LYL LG + R+ LS +
Sbjct 361 MSIVTMAMLSL-ACCVLESTVRATAVYWQE---LYLGMTLGMMRGVMGPMCRAILSHVAP 416
Query 88 EDELGKLFALLAIVEAVTPCVASLFYSILFAVSIDIYPGLVFMLAAS 134
E+GK+FAL +E+V+P A+ Y+ ++ +++ YPG ++A+
Sbjct 417 ATEVGKIFALTTSMESVSPLGAAPLYTTVYKATLENYPGAFNFISAA 463
> 7303930
Length=519
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 21/80 (26%), Positives = 46/80 (57%), Gaps = 4/80 (5%)
Query 54 AWQHVPGLYLSFILGSISPFSFIGVRSRLSKIINEDELGKLFALLAIVEAVTPCVASLFY 113
+WQ +Y++ LG R+ ++ ++ DE GK+FALL +++A++P ++S Y
Sbjct 415 SWQ----IYVAIGLGVFKSLVNPMCRTMITNLLPADERGKIFALLGVLQALSPLISSTLY 470
Query 114 SILFAVSIDIYPGLVFMLAA 133
++ +++ PG+ + +A
Sbjct 471 VAIYTRTLNTEPGIFNVFSA 490
> 7294102
Length=533
Score = 41.6 bits (96), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 32/130 (24%), Positives = 63/130 (48%), Gaps = 1/130 (0%)
Query 4 YSNVTSGMAVATLLLMGLLLPIFQNVFKFGDMRLALMGVLSLMAMCILRGAWQHVPGLYL 63
YSN + + A ++ M L +P+ + + D + +G + + + LY
Sbjct 352 YSNFKTFKSSAYVIAMLLAVPLMNKILGWRDTTIIFIGTWAHSIARLFFYFATNTDLLYA 411
Query 64 SFILGSISPFSFIGVRSRLSKIINEDELGKLFALLAIVEAVTPCVASLFYSILFAVSIDI 123
++ S+ P +R+ SKI+ E GK+FALL++ + P ++ + YS L+ + +
Sbjct 412 GAVVCSLGPIVGPMIRAMTSKIVPTSERGKVFALLSVCDNAVPFISGVCYSQLYRRTQNT 471
Query 124 -YPGLVFMLA 132
+ G VF+L
Sbjct 472 NHGGNVFILT 481
> CE21884
Length=475
Score = 41.2 bits (95), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 30/124 (24%), Positives = 60/124 (48%), Gaps = 2/124 (1%)
Query 9 SGMAVATLLLMGLLLPIFQNVFKFGDMRLALMGVLSL-MAMCILRGAWQHVPGLYLSFIL 67
M +M L L F D+RLA+ G+L+ + AW+ +++
Sbjct 283 KAMNTGMTTIMSLALYPFLKNLGITDIRLAIFGLLTRSIGRAWYAVAWEGWT-VFIVVFF 341
Query 68 GSISPFSFIGVRSRLSKIINEDELGKLFALLAIVEAVTPCVASLFYSILFAVSIDIYPGL 127
S F +RS ++ + E E G F+L+A++EA+ +S + + + +S++++P L
Sbjct 342 EMFSKFPATAMRSSIATNVGEHERGLAFSLVAVIEALCNLTSSWVFHLAWPLSLNVFPQL 401
Query 128 VFML 131
F++
Sbjct 402 SFVI 405
> Hs14780896
Length=475
Score = 38.5 bits (88), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 30/112 (26%), Positives = 60/112 (53%), Gaps = 8/112 (7%)
Query 25 IFQNVFKFGDMRLALMGVLSLMAMCILRGAWQHVPGLYLSFILGSISPFSFIGV---RSR 81
+F F+ D + ++G++S + +L V Y+ +I ++ F+ I V RS
Sbjct 340 VFSRCFR--DTTMIMIGMVSFGSGALLLA---FVKETYMFYIARAVMLFALIPVTTIRSA 394
Query 82 LSKIINEDELGKLFALLAIVEAVTPCVASLFYSILFAVSIDIYPGLVFMLAA 133
+SK+I GK+F +L + A+T V S Y+ ++ +++D++ G F L++
Sbjct 395 MSKLIKGSSYGKVFVILQLSLALTGVVTSTLYNKIYQLTMDMFVGSCFALSS 446
> CE27860
Length=434
Score = 38.5 bits (88), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 29/131 (22%), Positives = 62/131 (47%), Gaps = 20/131 (15%)
Query 4 YSNVTSGMAVATLLLMGLLLPIFQNVFKFGDMRLALMGVLSLMAMCIL-----RGAWQHV 58
Y++ S + ++ M + + F+ +F F D + + +LS M C+L + +W
Sbjct 256 YADYKSWRPIVQIVGMAVGMLFFKRIFHFRDTFIICLAILS-MTGCVLMIGLAQASW--- 311
Query 59 PGLYLSFILGSISPFSFIGVRSRLSK-----IINEDELGKLFALLAIVEAVTPCVASLFY 113
I S++P S G+ + +S I+ +DE+GK +A+ ++ + + SL
Sbjct 312 ------LIFASLAPGSLHGLLNPMSYTFIACIVEQDEIGKAYAISSVAQKLAGIAQSLVL 365
Query 114 SILFAVSIDIY 124
++ ++D Y
Sbjct 366 QNIYIATVDWY 376
> Hs18586019
Length=120
Score = 33.5 bits (75), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 36/74 (48%), Gaps = 5/74 (6%)
Query 52 RGAWQHVPGLYLSFILGSISPFSFIGVRSRLSKIINEDELGKLFALLAIVEAVTPCVASL 111
RG W+ GLYL +LG ++ S +G + + ++DEL +L ALL +PC
Sbjct 37 RGDWR---GLYL--LLGEVTGHSALGAQPAQAANEDDDELLELPALLQRPAGRSPCATLR 91
Query 112 FYSILFAVSIDIYP 125
I +I + P
Sbjct 92 GRRITPTTTILLLP 105
> Hs22035702
Length=234
Score = 30.8 bits (68), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 23/47 (48%), Gaps = 3/47 (6%)
Query 26 FQNVFKFGDMRLA-LMGVLSLMAMCILRGAWQHVPGLYLSFILGSIS 71
F+ F+ D LA L V+ M C L WQH P F+LGS S
Sbjct 98 FKTAFQLLDFILAVLWAVVWFMGFCFLANQWQHSPP--KEFLLGSSS 142
> HsM6912692
Length=234
Score = 30.8 bits (68), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 23/47 (48%), Gaps = 3/47 (6%)
Query 26 FQNVFKFGDMRLA-LMGVLSLMAMCILRGAWQHVPGLYLSFILGSIS 71
F+ F+ D LA L V+ M C L WQH P F+LGS S
Sbjct 98 FKTAFQLLDFILAVLWAVVWFMGFCFLANQWQHSPP--KEFLLGSSS 142
> CE00872
Length=341
Score = 30.0 bits (66), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 30/62 (48%), Gaps = 4/62 (6%)
Query 15 TLLLMGLLLPIFQNVFKFGDMRLALMGVLSLMAMCILRGAWQHVPGLYLSFILGSISPFS 74
+L+M LL P +++F F R + VL L I R + HV +FI I P +
Sbjct 85 NVLVMDLLGPSLEDLFNFCSRRFTMKTVLMLADQMIGRIEYVHVK----NFIHRDIKPDN 140
Query 75 FI 76
F+
Sbjct 141 FL 142
> 7302010
Length=388
Score = 27.3 bits (59), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 26/43 (60%), Gaps = 0/43 (0%)
Query 92 GKLFALLAIVEAVTPCVASLFYSILFAVSIDIYPGLVFMLAAS 134
GKLFA+ I+++ P VA+ Y+ ++ S+ PG L+A+
Sbjct 318 GKLFAIGNILQSFAPFVAAPLYTAIYKESLASNPGGFNFLSAA 360
Lambda K H
0.332 0.145 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1639544670
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40