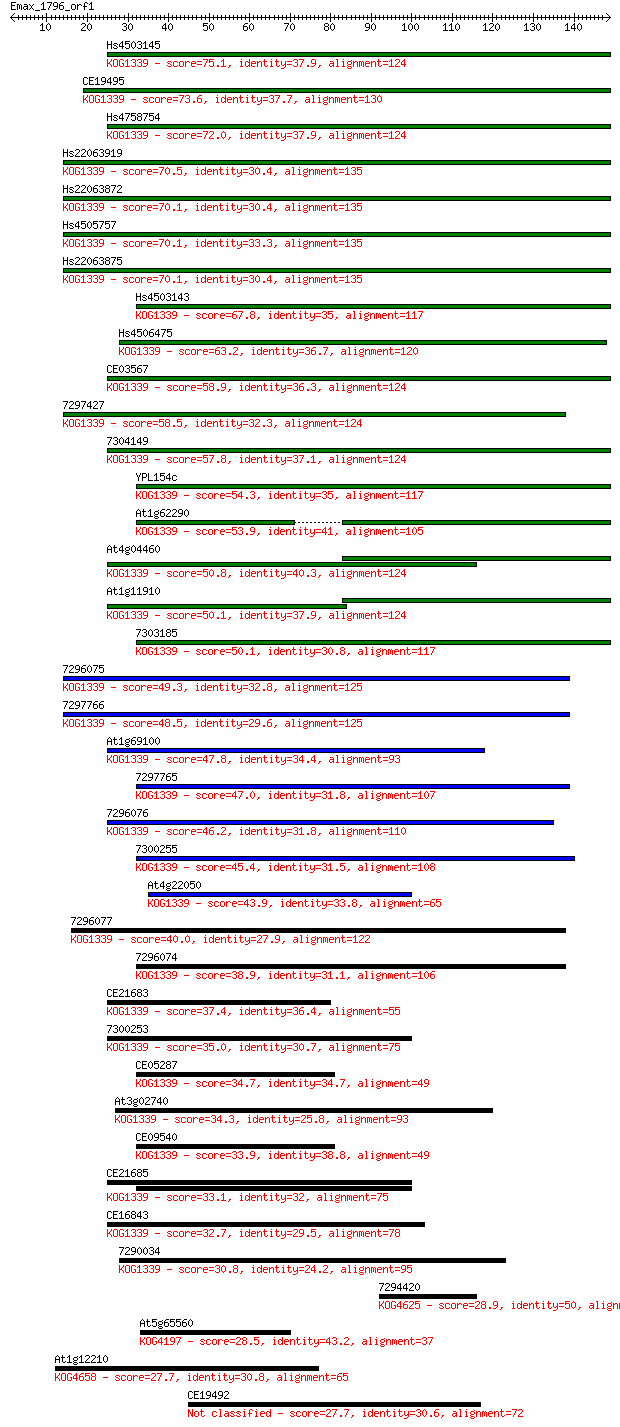
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1796_orf1
Length=148
Score E
Sequences producing significant alignments: (Bits) Value
Hs4503145 75.1 5e-14
CE19495 73.6 1e-13
Hs4758754 72.0 4e-13
Hs22063919 70.5 1e-12
Hs22063872 70.1 1e-12
Hs4505757 70.1 1e-12
Hs22063875 70.1 1e-12
Hs4503143 67.8 8e-12
Hs4506475 63.2 2e-10
CE03567 58.9 3e-09
7297427 58.5 4e-09
7304149 57.8 7e-09
YPL154c 54.3 9e-08
At1g62290 53.9 1e-07
At4g04460 50.8 1e-06
At1g11910 50.1 1e-06
7303185 50.1 2e-06
7296075 49.3 3e-06
7297766 48.5 5e-06
At1g69100 47.8 8e-06
7297765 47.0 1e-05
7296076 46.2 2e-05
7300255 45.4 3e-05
At4g22050 43.9 1e-04
7296077 40.0 0.002
7296074 38.9 0.003
CE21683 37.4 0.011
7300253 35.0 0.056
CE05287 34.7 0.059
At3g02740 34.3 0.089
CE09540 33.9 0.13
CE21685 33.1 0.18
CE16843 32.7 0.29
7290034 30.8 1.00
7294420 28.9 3.6
At5g65560 28.5 4.9
At1g12210 27.7 7.7
CE19492 27.7 7.9
> Hs4503145
Length=396
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 47/131 (35%), Positives = 69/131 (52%), Gaps = 17/131 (12%)
Query 25 PMTIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAE 84
P+T K YW+I L +++ G E GC+A VDTG+SLITGPS I L A+ A
Sbjct 249 PVT-KQAYWQIALDNIQVGGTVMFCSE--GCQAIVDTGTSLITGPSDKIKQLQNAIGAAP 305
Query 85 -------NCSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARGNPNNCAAGFMAM 137
C+NL +P +TF + V ++L+P Y + LD C++GF +
Sbjct 306 VDGEYAVECANLNVMPDVTFTING-----VPYTLSPTAYTL--LDFVDGMQFCSSGFQGL 358
Query 138 DVPAPRGPLFV 148
D+ P GPL++
Sbjct 359 DIHPPAGPLWI 369
> CE19495
Length=398
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 49/139 (35%), Positives = 72/139 (51%), Gaps = 21/139 (15%)
Query 19 NLLREPPMTIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVIN---- 74
N+ EP + DYW I L + I+G ++ + VDTG+SL+TGP+ VI
Sbjct 243 NIAWEP--LVSEDYWRIKLASVVIDGTTY---TSGPIDSIVDTGTSLLTGPTDVIKKIQH 297
Query 75 -----PLIKALNVAENCSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARGNPNN 129
PL E CS + +LP +TF L NF L +DY+++ + G +
Sbjct 298 KIGGIPLFNGEYEVE-CSKIPSLPNITFNLGG-----QNFDLQGKDYILQMSNGNGG-ST 350
Query 130 CAAGFMAMDVPAPRGPLFV 148
C +GFM MD+PAP GPL++
Sbjct 351 CLSGFMGMDIPAPAGPLWI 369
> Hs4758754
Length=420
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 47/132 (35%), Positives = 71/132 (53%), Gaps = 18/132 (13%)
Query 25 PMTIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALN--- 81
P+T+ YW+I + +K+ G +C K GC A +DTG+SLITGP+ I L A+
Sbjct 251 PVTVP-AYWQIHMERVKV-GPGLTLCAK-GCAAILDTGTSLITGPTEEIRALHAAIGGIP 307
Query 82 --VAEN---CSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARGNPNNCAAGFMA 136
E CS + LP ++F+L ++ F+L DYV++ R C +GF A
Sbjct 308 LLAGEYIILCSEIPKLPAVSFLLGGVW-----FNLTAHDYVIQT--TRNGVRLCLSGFQA 360
Query 137 MDVPAPRGPLFV 148
+DVP P GP ++
Sbjct 361 LDVPPPAGPFWI 372
> Hs22063919
Length=267
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 41/143 (28%), Positives = 75/143 (52%), Gaps = 22/143 (15%)
Query 14 ADLHTNLLREPPMTIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVI 73
+ +T L P+T++ YW+I + + +NG++ E GC+A VDTG+SL+TGP+S I
Sbjct 115 SSYYTGSLNWVPVTVE-GYWQITVDSITMNGEAIACAE--GCQAIVDTGTSLLTGPTSPI 171
Query 74 NPLIKALNVAEN--------CSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARG 125
+ + +EN CS + +LP + F + V + + P Y+++
Sbjct 172 ANIQSDIGASENSDGDMVVSCSAISSLPDIVFTING-----VQYPVPPSAYILQ------ 220
Query 126 NPNNCAAGFMAMDVPAPRGPLFV 148
+ +C +GF M++P G L++
Sbjct 221 SEGSCISGFQGMNLPTESGELWI 243
> Hs22063872
Length=325
Score = 70.1 bits (170), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 41/143 (28%), Positives = 75/143 (52%), Gaps = 22/143 (15%)
Query 14 ADLHTNLLREPPMTIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVI 73
+ +T L P+T++ YW+I + + +NG++ E GC+A VDTG+SL+TGP+S I
Sbjct 171 SSYYTGSLNWVPVTVE-GYWQITVDSITMNGEAIACAE--GCQAIVDTGTSLLTGPTSPI 227
Query 74 NPLIKALNVAEN--------CSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARG 125
+ + +EN CS + +LP + F + V + + P Y+++
Sbjct 228 ANIQSDIGASENSDGDMVVSCSAISSLPDIVFTING-----VQYPVPPSAYILQ------ 276
Query 126 NPNNCAAGFMAMDVPAPRGPLFV 148
+ +C +GF M++P G L++
Sbjct 277 SEGSCISGFQGMNLPTESGELWI 299
> Hs4505757
Length=388
Score = 70.1 bits (170), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 45/144 (31%), Positives = 74/144 (51%), Gaps = 22/144 (15%)
Query 14 ADLHTNLLREPPMTIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVI 73
+ L+T + P+T +L YW+IG+ I G++ G C + GC+A VDTG+SL+T P +
Sbjct 232 SSLYTGQIYWAPVTQEL-YWQIGIEEFLIGGQASGWCSE-GCQAIVDTGTSLLTVPQQYM 289
Query 74 NPLIKALNVAE--------NCSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARG 125
+ L++A E NC+++ LP+LTF++ V F L P Y++
Sbjct 290 SALLQATGAQEDEYGQFLVNCNSIQNLPSLTFIING-----VEFPLPPSSYILS------ 338
Query 126 NPNNCAAGFMAMDVPAPRG-PLFV 148
N C G + + G PL++
Sbjct 339 NNGYCTVGVEPTYLSSQNGQPLWI 362
> Hs22063875
Length=325
Score = 70.1 bits (170), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 41/143 (28%), Positives = 75/143 (52%), Gaps = 22/143 (15%)
Query 14 ADLHTNLLREPPMTIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVI 73
+ +T L P+T++ YW+I + + +NG++ E GC+A VDTG+SL+TGP+S I
Sbjct 171 SSYYTGSLNWVPVTVE-GYWQITVDSITMNGEAIACAE--GCQAIVDTGTSLLTGPTSPI 227
Query 74 NPLIKALNVAEN--------CSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARG 125
+ + +EN CS + +LP + F + V + + P Y+++
Sbjct 228 ANIQSDIGASENSDGDMVVSCSAISSLPDIVFTING-----VQYPVPPSAYILQ------ 276
Query 126 NPNNCAAGFMAMDVPAPRGPLFV 148
+ +C +GF M++P G L++
Sbjct 277 SEGSCISGFQGMNLPTESGELWI 299
> Hs4503143
Length=412
Score = 67.8 bits (164), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 41/125 (32%), Positives = 64/125 (51%), Gaps = 17/125 (13%)
Query 32 YWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAE------- 84
YW++ L +++ +C K GC A VDTG+SL+ GP + L KA+
Sbjct 269 YWQVHLDQVEV-ASGLTLC-KEGCEAIVDTGTSLMVGPVDEVRELQKAIGAVPLIQGEYM 326
Query 85 -NCSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARGNPNNCAAGFMAMDVPAPR 143
C + TLP +T L G+ + L+P DY ++ ++ C +GFM MD+P P
Sbjct 327 IPCEKVSTLPAITLKLG---GK--GYKLSPEDYTLKV--SQAGKTLCLSGFMGMDIPPPS 379
Query 144 GPLFV 148
GPL++
Sbjct 380 GPLWI 384
> Hs4506475
Length=406
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 44/127 (34%), Positives = 66/127 (51%), Gaps = 16/127 (12%)
Query 28 IKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAE--- 84
IK W+I + G+ + G S +CE GC A VDTG+S I+G +S I L++AL +
Sbjct 262 IKTGVWQIQMKGVSV-GSSTLLCED-GCLALVDTGASYISGSTSSIEKLMEALGAKKRLF 319
Query 85 ----NCSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARGNPNNCAAGFMAMDVP 140
C+ TLP ++F L G+ ++L DYV +E + + C AMD+P
Sbjct 320 DYVVKCNEGPTLPDISFHLG---GK--EYTLTSADYVFQE--SYSSKKLCTLAIHAMDIP 372
Query 141 APRGPLF 147
P GP +
Sbjct 373 PPTGPTW 379
> CE03567
Length=444
Score = 58.9 bits (141), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 45/133 (33%), Positives = 67/133 (50%), Gaps = 21/133 (15%)
Query 25 PMTIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVI---------NP 75
P+T K YW+ + K+ G C GC+A DTG+SLI GP + I P
Sbjct 267 PVTRK-GYWQFKMD--KVVGSGVLGCSN-GCQAIADTGTSLIAGPKAQIEAIQNFIGAEP 322
Query 76 LIKALNVAENCSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARGNPNNCAAGFM 135
LIK + +C + TLP ++FV+ FSL DYV++ ++G C +GFM
Sbjct 323 LIKGEYMI-SCDKVPTLPPVSFVIGG-----QEFSLKGEDYVLKV--SQGGKTICLSGFM 374
Query 136 AMDVPAPRGPLFV 148
+D+P G L++
Sbjct 375 GIDLPERVGELWI 387
> 7297427
Length=372
Score = 58.5 bits (140), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 40/128 (31%), Positives = 65/128 (50%), Gaps = 20/128 (15%)
Query 14 ADLHTNLLREPPMTIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVI 73
A L++ L P++ + YW+ + G I+G S +C+ C+A DTG+SLI P +
Sbjct 229 ASLYSGALTYVPIS-EQGYWQFTMAGSSIDGYS--LCDD--CQAIADTGTSLIVAPYNAY 283
Query 74 NPLIKALNVAE----NCSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARGNPNN 129
L + LNV E +CS++ +LP +TF + NF L P Y+++ + N
Sbjct 284 ITLSEILNVGEDGYLDCSSVSSLPDVTFNIGG-----TNFVLKPSAYIIQ------SDGN 332
Query 130 CAAGFMAM 137
C + F M
Sbjct 333 CMSAFEYM 340
> 7304149
Length=392
Score = 57.8 bits (138), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 46/135 (34%), Positives = 69/135 (51%), Gaps = 25/135 (18%)
Query 25 PMTIKLDYWEIGLHGMKINGKSFGVCE--KRGCRAAVDTGSSLITGP---SSVIN----- 74
P+T K YW+I K++ S G + K GC+ DTG+SLI P ++ IN
Sbjct 246 PVTRKA-YWQI-----KMDAASIGDLQLCKGGCQVIADTGTSLIAAPLEEATSINQKIGG 299
Query 75 -PLIKALNVAENCSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARGNPNNCAAG 133
P+I V +C + LP + FVL G+ F L +DY++ A+ C +G
Sbjct 300 TPIIGGQYVV-SCDLIPQLPVIKFVLG---GK--TFELEGKDYILRV--AQMGKTICLSG 351
Query 134 FMAMDVPAPRGPLFV 148
FM +D+P P GPL++
Sbjct 352 FMGLDIPPPNGPLWI 366
> YPL154c
Length=405
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 41/125 (32%), Positives = 56/125 (44%), Gaps = 22/125 (17%)
Query 32 YWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPS---SVINPLIKAL-----NVA 83
YWE+ G+ + G + E G AA+DTG+SLIT PS +IN I A
Sbjct 269 YWEVKFEGIGL-GDEYAELESHG--AAIDTGTSLITLPSGLAEMINAEIGAKKGWTGQYT 325
Query 84 ENCSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARGNPNNCAAGFMAMDVPAPR 143
+C+ LP L F NF++ P DY +E +C + MD P P
Sbjct 326 LDCNTRDNLPDLIFNFNG-----YNFTIGPYDYTLEV------SGSCISAITPMDFPEPV 374
Query 144 GPLFV 148
GPL +
Sbjct 375 GPLAI 379
> At1g62290
Length=526
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 27/66 (40%), Positives = 41/66 (62%), Gaps = 7/66 (10%)
Query 83 AENCSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARGNPNNCAAGFMAMDVPAP 142
A +CS L +PT++F I G++ F LAP +YV++ G C +GF A+D+P P
Sbjct 442 AVDCSQLSKMPTVSFT---IGGKV--FDLAPEEYVLK--IGEGPVAQCISGFTALDIPPP 494
Query 143 RGPLFV 148
RGPL++
Sbjct 495 RGPLWI 500
Score = 37.0 bits (84), Expect = 0.013, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 24/39 (61%), Gaps = 1/39 (2%)
Query 32 YWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPS 70
YW+ + + I G+S G C GC A D+G+SL+ GP+
Sbjct 267 YWQFDMGEVLIAGESTGYC-GSGCSAIADSGTSLLAGPT 304
> At4g04460
Length=508
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 25/66 (37%), Positives = 39/66 (59%), Gaps = 7/66 (10%)
Query 83 AENCSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARGNPNNCAAGFMAMDVPAP 142
A +C + ++P +TF I GR +F L P+DY+ + G + C +GF AMD+ P
Sbjct 424 AVDCGRVSSMPIVTF---SIGGR--SFDLTPQDYIFK--IGEGVESQCTSGFTAMDIAPP 476
Query 143 RGPLFV 148
RGPL++
Sbjct 477 RGPLWI 482
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 33/92 (35%), Positives = 49/92 (53%), Gaps = 9/92 (9%)
Query 25 PMTIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAE 84
P+T K YW+ + ++I GK G C K GC A D+G+SL+TGPS+VI + A+
Sbjct 259 PVTHK-GYWQFDMGDLQIAGKPTGYCAK-GCSAIADSGTSLLTGPSTVITMINHAIGAQ- 315
Query 85 NCSNLGTLPTLTFVLKDIYGR-LVNFSLAPRD 115
G + + D YG+ ++N LA D
Sbjct 316 -----GIVSRECKAVVDQYGKTMLNSLLAQED 342
> At1g11910
Length=506
Score = 50.1 bits (118), Expect = 1e-06, Method: Composition-based stats.
Identities = 26/66 (39%), Positives = 41/66 (62%), Gaps = 7/66 (10%)
Query 83 AENCSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARGNPNNCAAGFMAMDVPAP 142
A +C+ L T+PT++ I G++ F LAP +YV++ G C +GF+A+DV P
Sbjct 422 AVDCAQLSTMPTVSLT---IGGKV--FDLAPEEYVLKV--GEGPVAQCISGFIALDVAPP 474
Query 143 RGPLFV 148
RGPL++
Sbjct 475 RGPLWI 480
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 34/59 (57%), Gaps = 2/59 (3%)
Query 25 PMTIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVA 83
P+T K YW+ + + I G G CE GC A D+G+SL+ GP+++I + A+ A
Sbjct 254 PVTQK-GYWQFDMGDVLIGGAPTGFCES-GCSAIADSGTSLLAGPTTIITMINHAIGAA 310
> 7303185
Length=404
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 36/125 (28%), Positives = 63/125 (50%), Gaps = 18/125 (14%)
Query 32 YWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGP---SSVINPLIKALNVAEN--- 85
YW++ + I ++ +C++ GC +DTG+S + P + +IN I +
Sbjct 263 YWQVKMDSAVI--RNLELCQQ-GCEVIIDTGTSFLALPYDQAILINESIGGTPSSFGQFL 319
Query 86 --CSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARGNPNNCAAGFMAMDVPAPR 143
C ++ LP +TF L GR F L +YV D + C++ F+A+D+P+P
Sbjct 320 VPCDSVPDLPKITFTLG---GR--RFFLESHEYVFR--DIYQDRRICSSAFIAVDLPSPS 372
Query 144 GPLFV 148
GPL++
Sbjct 373 GPLWI 377
> 7296075
Length=410
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 41/135 (30%), Positives = 61/135 (45%), Gaps = 26/135 (19%)
Query 14 ADLHTNLLREPPMTIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSV- 72
+ L++ L P++ YW+ + +NG F CE C A +D G+SLI P V
Sbjct 259 SGLYSGCLTYVPVS-SAGYWQFTMTSANLNG--FQFCEN--CEAILDVGTSLIVVPEQVL 313
Query 73 --INPLIKALN-VAEN------CSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDA 123
IN ++ LN A N CS++G LP + F + F L DYV+
Sbjct 314 DTINQILGVLNPTASNGVFLVDCSSIGDLPDIVFTVAR-----RKFPLKSSDYVLRY--- 365
Query 124 RGNPNNCAAGFMAMD 138
N C +GF +M+
Sbjct 366 ---GNTCVSGFTSMN 377
> 7297766
Length=391
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 37/132 (28%), Positives = 64/132 (48%), Gaps = 19/132 (14%)
Query 14 ADLHTNLLREPPMTIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVI 73
+ L+ L P+++ YW+ ++ +K NG +C GC+A DTG+SLI P +
Sbjct 238 SSLYRGSLTYVPVSVPA-YWQFKVNTIKTNGTL--LCN--GCQAIADTGTSLIAVPLAAY 292
Query 74 NPLIKALNVAEN-------CSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARGN 126
+ + L +N C + +LP + +I G + F+LAPRDY+V+ +
Sbjct 293 RKINRQLGATDNDGEAFVRCGRVSSLPKVNL---NIGGTV--FTLAPRDYIVKV--TQNG 345
Query 127 PNNCAAGFMAMD 138
C + F M+
Sbjct 346 QTYCMSAFTYME 357
> At1g69100
Length=343
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 32/97 (32%), Positives = 46/97 (47%), Gaps = 9/97 (9%)
Query 25 PMTIKLDYWEIGLHGMKINGK-SFGVCEKRGCRAAVDTGSSLITGPSSVINPLIK---AL 80
PM + D W+I + + INGK + C+ C A VD+GS+ I GP + + K A
Sbjct 198 PMKLSDDRWKIKMSKIYINGKPAINFCDDVECTAMVDSGSTDIFGPDEAVGKIYKEIGAT 257
Query 81 NVAENCSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYV 117
V C LP + F +I G+ + L DYV
Sbjct 258 KVIIRCEQFPALPDIYF---EIGGK--HLRLTKHDYV 289
> 7297765
Length=423
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 34/115 (29%), Positives = 54/115 (46%), Gaps = 18/115 (15%)
Query 32 YWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAE------- 84
YW+ + +KI G + GC+A DTG+SLI P + K LN +
Sbjct 252 YWQFTANSVKIEG----ILLCNGCQAIADTGTSLIAVPLRAYKAINKVLNATDAGDGEAF 307
Query 85 -NCSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARGNPNNCAAGFMAMD 138
+CS+L LP + +I G ++L P+DY+ ++ A N C +GF +
Sbjct 308 VDCSSLCRLPNVNL---NIGG--TTYTLTPKDYIY-KVQADNNQTLCLSGFTYLQ 356
> 7296076
Length=405
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 35/117 (29%), Positives = 54/117 (46%), Gaps = 19/117 (16%)
Query 25 PMTIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSV---INPLIKALN 81
P+T + YW+ ++ + NG GC+A DTG+SLI PS+ +N LI +
Sbjct 266 PVT-QQGYWQFAVNNITWNGTVI----SSGCQAIADTGTSLIAAPSAAYIQLNNLIGGVP 320
Query 82 VAEN----CSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARGNPNNCAAGF 134
+ + CS + +LP LT + NF L P Y+ + GN C + F
Sbjct 321 IQGDYYVPCSTVSSLPVLTINIGG-----TNFYLPPSVYI--QTYTEGNYTTCMSTF 370
> 7300255
Length=395
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 34/116 (29%), Positives = 56/116 (48%), Gaps = 18/116 (15%)
Query 32 YWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSS---VINPLIKALNVAEN--- 85
YW+ L +++ G + + R +A DTG+SL+ P +IN L+ L + N
Sbjct 260 YWQFPLDVIEVAGTR--INQNR--QAIADTGTSLLAAPPREYLIINSLLGGLPTSNNEYL 315
Query 86 --CSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARGNPNNCAAGFMAMDV 139
CS + +LP + F++ F L PRDYV+ + G+ + C + F MD
Sbjct 316 LNCSEIDSLPEIVFIIGG-----QRFGLQPRDYVMSATNDDGS-SICLSAFTLMDA 365
> At4g22050
Length=336
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 36/65 (55%), Gaps = 1/65 (1%)
Query 35 IGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAENCSNLGTLPT 94
+ + + GK+ +C GC+A VD+GSS I P + + + + V NC+N TLP
Sbjct 227 FAMSNIWVGGKNTNICSS-GCKAIVDSGSSNINVPMDSADEIHRYIGVEPNCNNFETLPD 285
Query 95 LTFVL 99
+TF +
Sbjct 286 VTFTI 290
> 7296077
Length=418
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 34/132 (25%), Positives = 61/132 (46%), Gaps = 26/132 (19%)
Query 16 LHTNLLREPPMTIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINP 75
L++ L P++ K+ +W+I + ++I K +C C+A D G+SLI P +
Sbjct 257 LYSGCLTYVPVS-KVGFWQITVGQVEIGSKK--LCSN--CQAIFDMGTSLIIVPCPALKI 311
Query 76 LIKALNVAE----------NCSNLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARG 125
+ K L + E +C + LP + F + +F+L P DY+ L+ G
Sbjct 312 INKKLGIKETDRKDGVYIIDCKKVSHLPKIVFNIG-----WKDFTLNPSDYI---LNYSG 363
Query 126 NPNNCAAGFMAM 137
C +GF ++
Sbjct 364 ---TCVSGFSSL 372
> 7296074
Length=393
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 33/112 (29%), Positives = 51/112 (45%), Gaps = 15/112 (13%)
Query 32 YWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAENCS-NLG 90
YW L + ++GK + G +A +DTG+SLI GP + + K + N + NL
Sbjct 258 YWSFKLDFIAVHGKG-SRSSRTGNKAIMDTGTSLIVGPVLEVLYINKDIGAEHNKTYNLY 316
Query 91 T-----LPTLTFVLKDIYGRLVNFSLAPRDYVVEELDARGNPNNCAAGFMAM 137
T +P L ++ I G+ F + P YV+ N C + FM M
Sbjct 317 TVACESIPQLPIIVFGIAGK--EFFVKPHTYVIRY------DNFCFSAFMDM 360
> CE21683
Length=320
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 29/55 (52%), Gaps = 5/55 (9%)
Query 25 PMTIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKA 79
P+T K YWE L G + S E + + DTG+S I P S+I+ ++KA
Sbjct 247 PLTAKT-YWEFALDGFAVGSFS----ETKTAKVISDTGTSWIGAPHSIISAVVKA 296
> 7300253
Length=465
Score = 35.0 bits (79), Expect = 0.056, Method: Composition-based stats.
Identities = 23/81 (28%), Positives = 37/81 (45%), Gaps = 11/81 (13%)
Query 25 PMTIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAE 84
P+T K YW+ L + + G +A VD+G+SLIT P+++ N + K +
Sbjct 323 PVT-KKGYWQFTLQDIYVGGTKV----SGSVQAIVDSGTSLITAPTAIYNKINKVIGCRA 377
Query 85 NCSN------LGTLPTLTFVL 99
S +P TFV+
Sbjct 378 TSSGECWMKCAKKIPDFTFVI 398
> CE05287
Length=428
Score = 34.7 bits (78), Expect = 0.059, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 26/49 (53%), Gaps = 2/49 (4%)
Query 32 YWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKAL 80
+W+ L G ++ S+ G +AA DT +S I P SV+ L KA+
Sbjct 292 FWQFKLGG--VSSGSYSQAPNSGWQAAADTAASFIGAPKSVVTSLAKAV 338
> At3g02740
Length=488
Score = 34.3 bits (77), Expect = 0.089, Method: Composition-based stats.
Identities = 24/109 (22%), Positives = 54/109 (49%), Gaps = 22/109 (20%)
Query 27 TIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKAL------ 80
++ L+ E+G ++++ +F + +G +D+G++L+ P +V NPL+ +
Sbjct 285 SVNLNAIEVGNSVLELSSNAFDSGDDKG--VIIDSGTTLVYLPDAVYNPLLNEILASHPE 342
Query 81 ----NVAENCS------NLGTLPTLTFVLKDIYGRLVNFSLAPRDYVVE 119
V E+ + L PT+TF + + V+ ++ PR+Y+ +
Sbjct 343 LTLHTVQESFTCFHYTDKLDRFPTVTFQ----FDKSVSLAVYPREYLFQ 387
> CE09540
Length=395
Score = 33.9 bits (76), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 26/49 (53%), Gaps = 4/49 (8%)
Query 32 YWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKAL 80
Y++ G K+ S + + DTGSSLI GPS+VIN +AL
Sbjct 255 YYQFVADGFKLGSYS----NNKKSQVISDTGSSLIGGPSAVINGFAQAL 299
> CE21685
Length=829
Score = 33.1 bits (74), Expect = 0.18, Method: Composition-based stats.
Identities = 24/85 (28%), Positives = 39/85 (45%), Gaps = 16/85 (18%)
Query 25 PMTIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAV-DTGSSLITGPSSVINPLIKALNVA 83
P+T K YW+ K++G + G + G + DTGSS I+ P ++IN + +
Sbjct 682 PLTAK-TYWQ-----FKMDGFAVGTYSQYGYNQVISDTGSSWISAPYAMINDIATQTHAT 735
Query 84 EN---------CSNLGTLPTLTFVL 99
+ CS + T P L F +
Sbjct 736 WDEMNEIYTVKCSTMKTQPDLVFTI 760
Score = 29.6 bits (65), Expect = 2.0, Method: Composition-based stats.
Identities = 19/77 (24%), Positives = 34/77 (44%), Gaps = 13/77 (16%)
Query 32 YWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAE------- 84
YW+ + G+ I + E+ A DTGS+ + P+ V+N +++
Sbjct 254 YWQFPMDGVAIGNYAMMKQEQ----AISDTGSAWLGLPNPVLNAIVQQTKATYDWNYEIY 309
Query 85 --NCSNLGTLPTLTFVL 99
+CS + T P L F +
Sbjct 310 TLDCSTMQTQPDLVFTI 326
> CE16843
Length=394
Score = 32.7 bits (73), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 23/87 (26%), Positives = 36/87 (41%), Gaps = 13/87 (14%)
Query 25 PMTIKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAE 84
P+T K YW+ + G+ I+ + K G DTG+S I GP VI +
Sbjct 249 PLT-KAAYWQFRMQGIGIDNVN---EHKNGWEVISDTGTSFIGGPGKVIQDIANKYGATY 304
Query 85 N---------CSNLGTLPTLTFVLKDI 102
+ CS + L +L + D+
Sbjct 305 DEYSDSYVIPCSEIKNLRSLKMKINDL 331
> 7290034
Length=407
Score = 30.8 bits (68), Expect = 1.00, Method: Compositional matrix adjust.
Identities = 23/97 (23%), Positives = 45/97 (46%), Gaps = 10/97 (10%)
Query 28 IKLDYWEIGLHGMKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAENCS 87
++ YW+ +K+ S +A DTG+SLI P + + + + N N
Sbjct 251 VQAAYWKFQTDYIKVGSTSISTF----AQAIADTGTSLIIAPQAQYDQISQLFNA--NSE 304
Query 88 NLGTLPTLTF--VLKDIYGRLVNFSLAPRDYVVEELD 122
L + ++ ++ +I G V+F + + Y++EE D
Sbjct 305 GLFECSSTSYPDLIINING--VDFKIPAKYYIIEEED 339
> 7294420
Length=1780
Score = 28.9 bits (63), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 12/24 (50%), Positives = 15/24 (62%), Gaps = 0/24 (0%)
Query 92 LPTLTFVLKDIYGRLVNFSLAPRD 115
+P + L D+YGR V SL PRD
Sbjct 148 MPKPLWALVDLYGRCVQVSLCPRD 171
> At5g65560
Length=915
Score = 28.5 bits (62), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 22/43 (51%), Gaps = 6/43 (13%)
Query 33 WEIGLHGMKING------KSFGVCEKRGCRAAVDTGSSLITGP 69
W+I + G+ G + F V EK GC+ + T S LI GP
Sbjct 869 WKIIIDGVGKQGLVEAFYELFNVMEKNGCKFSSQTYSLLIEGP 911
> At1g12210
Length=885
Score = 27.7 bits (60), Expect = 7.7, Method: Composition-based stats.
Identities = 20/68 (29%), Positives = 30/68 (44%), Gaps = 9/68 (13%)
Query 12 KTADLHTNLLREPPMTIKLDYWEIGLHGMKINGKSFGV---CEKRGCRAAVDTGSSLITG 68
+ D+H L R+ + + D WE K+ K GV + GC+ A T S + G
Sbjct 247 RALDIHNVLRRKKFVLLLDDIWE------KVELKVIGVPYPSGENGCKVAFTTHSKEVCG 300
Query 69 PSSVINPL 76
V NP+
Sbjct 301 RMGVDNPM 308
> CE19492
Length=481
Score = 27.7 bits (60), Expect = 7.9, Method: Composition-based stats.
Identities = 22/75 (29%), Positives = 34/75 (45%), Gaps = 11/75 (14%)
Query 45 KSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAENCSNLGTLPTLTFVLKDIYG 104
K G E C+A DTG+ PSS N LI + A + L P + ++++
Sbjct 285 KKMGTLENENCKAYADTGT-----PSSPSNTLIDFKSDAYSLPQLFPYPYFSIIVENCD- 338
Query 105 RLVNFSLA---PRDY 116
VNF+ + P +Y
Sbjct 339 --VNFNFSTTLPSNY 351
Lambda K H
0.319 0.138 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1821716300
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40