bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1774_orf1
Length=133
Score E
Sequences producing significant alignments: (Bits) Value
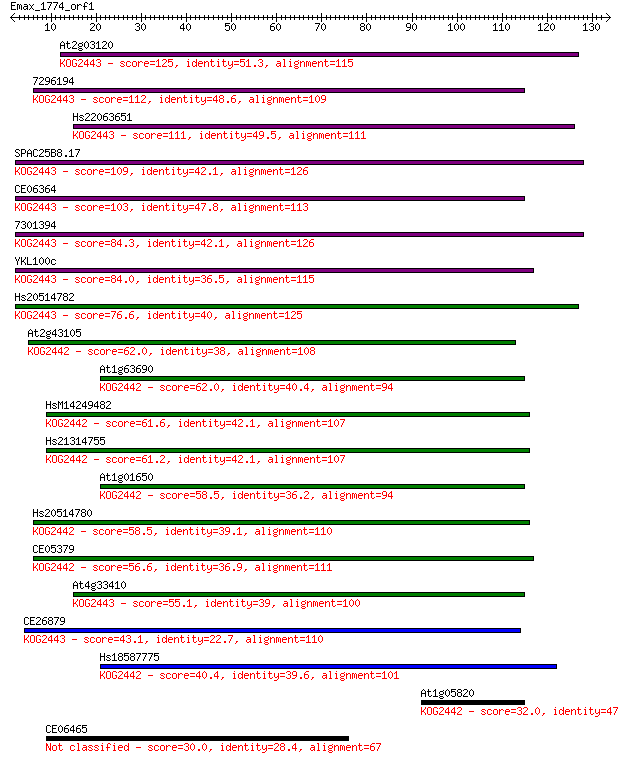
At2g03120 125 2e-29
7296194 112 2e-25
Hs22063651 111 5e-25
SPAC25B8.17 109 1e-24
CE06364 103 8e-23
7301394 84.3 6e-17
YKL100c 84.0 6e-17
Hs20514782 76.6 1e-14
At2g43105 62.0 3e-10
At1g63690 62.0 3e-10
HsM14249482 61.6 4e-10
Hs21314755 61.2 5e-10
At1g01650 58.5 3e-09
Hs20514780 58.5 4e-09
CE05379 56.6 1e-08
At4g33410 55.1 3e-08
CE26879 43.1 1e-04
Hs18587775 40.4 0.001
At1g05820 32.0 0.36
CE06465 30.0 1.3
> At2g03120
Length=344
Score = 125 bits (315), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 59/115 (51%), Positives = 80/115 (69%), Gaps = 0/115 (0%)
Query 12 CTLWLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVFGTEVMVSV 71
C + KHW +NIL ++FCIQ I ++S+GSF +ILL GLF YD+ WVF T VMVSV
Sbjct 152 CAWYAWKKHWLANNILGLSFCIQGIEMLSLGSFKTGAILLAGLFFYDIFWVFFTPVMVSV 211
Query 72 AKAFEGPAKLMFPVQISPLQYSILGLGDVVIPGVLIAMCLRFDLFLYLKQQNATA 126
AK+F+ P KL+FP + YS+LGLGD+VIPG+ +A+ LRFD+ + Q T+
Sbjct 212 AKSFDAPIKLLFPTGDALRPYSMLGLGDIVIPGIFVALALRFDVSRRRQPQYFTS 266
> 7296194
Length=389
Score = 112 bits (280), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 53/114 (46%), Positives = 77/114 (67%), Gaps = 5/114 (4%)
Query 6 LVALAICTLWLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVFGT 65
+++ AI +LL KHW +N+ +AF I + ++ + +F ILL GLF YD+ WVFGT
Sbjct 176 VISSAIGVWYLLKKHWIANNLFGLAFAINGVEMLHLNNFVTGVILLSGLFFYDIFWVFGT 235
Query 66 EVMVSVAKAFEGPAKLMFPVQ-----ISPLQYSILGLGDVVIPGVLIAMCLRFD 114
VMV+VAK+FE P KL+FP ++ +++LGLGD+VIPG+ IA+ LRFD
Sbjct 236 NVMVTVAKSFEAPIKLVFPQDLIENGLNASNFAMLGLGDIVIPGIFIALLLRFD 289
> Hs22063651
Length=377
Score = 111 bits (277), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 55/116 (47%), Positives = 76/116 (65%), Gaps = 7/116 (6%)
Query 15 WLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVFGTEVMVSVAKA 74
+LL KHW +N+ +AF + + L+ + + ILL GLF+YDV WVFGT VMV+VAK+
Sbjct 176 YLLRKHWIANNLFGLAFSLNGVELLHLNNVSTGCILLGGLFIYDVFWVFGTNVMVTVAKS 235
Query 75 FEGPAKLMFPVQ-----ISPLQYSILGLGDVVIPGVLIAMCLRFDLFLYLKQQNAT 125
FE P KL+FP + +++LGLGDVVIPG+ IA+ LRFD + LK+ T
Sbjct 236 FEAPIKLVFPQDLLEKGLEANNFAMLGLGDVVIPGIFIALLLRFD--ISLKKNTHT 289
> SPAC25B8.17
Length=295
Score = 109 bits (273), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 53/126 (42%), Positives = 83/126 (65%), Gaps = 3/126 (2%)
Query 2 IISHLVALAICTLWLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIW 61
I + + ++AI + TKHW NILA A +I+++ + S+ ++LL LF YD+ +
Sbjct 93 ITATMSSIAIALFYFKTKHWMASNILAWALAANSISIMRIDSYNTGALLLGALFFYDIYF 152
Query 62 VFGTEVMVSVAKAFEGPAKLMFPVQISPLQYSILGLGDVVIPGVLIAMCLRFDLFLYLKQ 121
VFGTEVMV+VA + PAK + P +P + S+LGLGD+V+PG+++A+ RFDL Y+
Sbjct 153 VFGTEVMVTVATGIDIPAKYVLPQFKNPTRLSMLGLGDIVMPGLMLALMYRFDLHYYI-- 210
Query 122 QNATAQ 127
N+T+Q
Sbjct 211 -NSTSQ 215
> CE06364
Length=468
Score = 103 bits (257), Expect = 8e-23, Method: Composition-based stats.
Identities = 54/119 (45%), Positives = 73/119 (61%), Gaps = 6/119 (5%)
Query 2 IISHLVALAICTLWLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIW 61
II+ L+ I LL +HW +NI+ ++F I I + + SF S+LL GLF YD+ W
Sbjct 250 IIALLMCSPILISHLLKRHWITNNIIGVSFSILGIERLHLASFKAGSLLLVGLFFYDIFW 309
Query 62 VFGTEVMVSVAKAFEGPAKLMFPVQI------SPLQYSILGLGDVVIPGVLIAMCLRFD 114
VFGT+VM SVAK + P L FP I ++S+LGLGD+VIPG+ IA+ RFD
Sbjct 310 VFGTDVMTSVAKGIDAPILLQFPQDIYRNGIMEASKHSMLGLGDIVIPGIFIALLRRFD 368
> 7301394
Length=417
Score = 84.3 bits (207), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 53/156 (33%), Positives = 78/156 (50%), Gaps = 32/156 (20%)
Query 2 IISHLVALAICTLWLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIW 61
+ S ++++I +W+LT HW L + + + C+ IA V + S V+++LL GL +YDV W
Sbjct 180 LFSFTLSVSIVCIWVLTGHWLLMDAMGMGLCVAFIAFVRLPSLKVSTLLLTGLLIYDVFW 239
Query 62 V------FGTEVMVSVA------------------------KAFEGPAKLMFPVQISPLQ 91
V F T VMV VA P KL+FP +
Sbjct 240 VFLSSYIFSTNVMVKVATRPADNPVGIVARKLHLGGIVRDTPKLNLPGKLVFPSLHNTGH 299
Query 92 YSILGLGDVVIPGVLIAMCLRFDLFLYLKQQNATAQ 127
+S+LGLGDVV+PG+L+ LR+D Y K Q T+
Sbjct 300 FSMLGLGDVVMPGLLLCFVLRYD--AYKKAQGVTSD 333
> YKL100c
Length=587
Score = 84.0 bits (206), Expect = 6e-17, Method: Composition-based stats.
Identities = 42/120 (35%), Positives = 76/120 (63%), Gaps = 5/120 (4%)
Query 2 IISHLVALAICTLWLLTKH-WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVI 60
I+S ++++ + L+ + W + N +++ I +IA + + + +++L LF YD+
Sbjct 309 IVSFVLSIVSTVYFYLSPNDWLISNAVSMNMAIWSIAQLKLKNLKSGALILIALFFYDIC 368
Query 61 WVFGTEVMVSVAKAFEGPAKLMFPVQISPLQ----YSILGLGDVVIPGVLIAMCLRFDLF 116
+VFGT+VMV+VA + P KL PV+ + Q +SILGLGD+ +PG+ IAMC ++D++
Sbjct 369 FVFGTDVMVTVATNLDIPVKLSLPVKFNTAQNNFNFSILGLGDIALPGMFIAMCYKYDIW 428
> Hs20514782
Length=385
Score = 76.6 bits (187), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 50/156 (32%), Positives = 77/156 (49%), Gaps = 34/156 (21%)
Query 2 IISHLVALAICTLWLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIW 61
++S +++ + +W+LT HW L + LA+ C+ IA V + S V+ +LL GL +YDV W
Sbjct 145 LLSFSLSVMLVLIWVLTGHWLLMDALAMGLCVAMIAFVRLPSLKVSCLLLSGLLIYDVFW 204
Query 62 V------FGTEVMVSVA-------------------------KAFEGPAKLMFPVQISPL 90
V F + VMV VA P KL+FP
Sbjct 205 VFFSAYIFNSNVMVKVATQPADNPLDVLSRKLHLGPNVGRDVPRLSLPGKLVFPSSTGS- 263
Query 91 QYSILGLGDVVIPGVLIAMCLRFDLFLYLKQQNATA 126
+S+LG+GD+V+PG+L+ LR+D Y KQ + +
Sbjct 264 HFSMLGIGDIVMPGLLLCFVLRYD--NYKKQASGDS 297
> At2g43105
Length=543
Score = 62.0 bits (149), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 41/124 (33%), Positives = 61/124 (49%), Gaps = 16/124 (12%)
Query 5 HLVALAICTLWLLTKH----WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVI 60
++V LA W + +H W +IL I I A+ +V + + VA++LLC FVYD+
Sbjct 330 NIVCLAFAVFWFIKRHTSYSWVGQDILGICLMITALQVVRLPNIKVATVLLCCAFVYDIF 389
Query 61 WV------FGTEVMVSVAKAFEG-----PAKLMFPVQISPL-QYSILGLGDVVIPGVLIA 108
WV F VM+ VA+ P L P P Y ++G GD++ PG+LI+
Sbjct 390 WVFISPLIFHESVMIVVAQGDSSTGESIPMLLRIPRFFDPWGGYDMIGFGDILFPGLLIS 449
Query 109 MCLR 112
R
Sbjct 450 FASR 453
> At1g63690
Length=519
Score = 62.0 bits (149), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 38/106 (35%), Positives = 58/106 (54%), Gaps = 12/106 (11%)
Query 21 WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVFGTE------VMVSVAKA 74
W ++L IA I + +V V + V ++LL F+YD+ WVF ++ VM+ VA+
Sbjct 348 WIGQDVLGIALIITVLQIVHVPNLKVGTVLLSCAFLYDIFWVFVSKKLFHESVMIVVARG 407
Query 75 FEG-----PAKLMFPVQISPL-QYSILGLGDVVIPGVLIAMCLRFD 114
+ P L P P YSI+G GD+++PG+LIA LR+D
Sbjct 408 DKSGEDGIPMLLKIPRMFDPWGGYSIIGFGDILLPGLLIAFALRYD 453
> HsM14249482
Length=409
Score = 61.6 bits (148), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 45/132 (34%), Positives = 68/132 (51%), Gaps = 25/132 (18%)
Query 9 LAICTLWLLTKH-----WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVF 63
+A+ +W + ++ W L +IL IAFC+ I + + +F ILL L +YDV +VF
Sbjct 186 IAVAVVWAVFRNEDRWAWILQDILGIAFCLNLIKTLKLPNFKSCVILLGLLLLYDVFFVF 245
Query 64 --------GTEVMVSVAKA-FEGPAKLMFPVQISPLQY-----------SILGLGDVVIP 103
G +MV +A F KL +++ L Y SILG GD+++P
Sbjct 246 ITPFITKNGESIMVELAAGPFGNNEKLPVVIRVPKLIYFSVMSVCLMPVSILGFGDIIVP 305
Query 104 GVLIAMCLRFDL 115
G+LIA C RFD+
Sbjct 306 GLLIAYCRRFDV 317
> Hs21314755
Length=520
Score = 61.2 bits (147), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 45/132 (34%), Positives = 68/132 (51%), Gaps = 25/132 (18%)
Query 9 LAICTLWLLTKH-----WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVF 63
+A+ +W + ++ W L +IL IAFC+ I + + +F ILL L +YDV +VF
Sbjct 297 IAVAVVWAVFRNEDRWAWILQDILGIAFCLNLIKTLKLPNFKSCVILLGLLLLYDVFFVF 356
Query 64 --------GTEVMVSVAKA-FEGPAKLMFPVQISPLQY-----------SILGLGDVVIP 103
G +MV +A F KL +++ L Y SILG GD+++P
Sbjct 357 ITPFITKNGESIMVELAAGPFGNNEKLPVVIRVPKLIYFSVMSVCLMPVSILGFGDIIVP 416
Query 104 GVLIAMCLRFDL 115
G+LIA C RFD+
Sbjct 417 GLLIAYCRRFDV 428
> At1g01650
Length=491
Score = 58.5 bits (140), Expect = 3e-09, Method: Composition-based stats.
Identities = 34/106 (32%), Positives = 55/106 (51%), Gaps = 12/106 (11%)
Query 21 WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVFGTE------VMVSVAKA 74
+ L ++ I+ I + +V V + V +LL F+YD+ WVF ++ VM+ VA+
Sbjct 301 FGLSTLMGISLIITVLQIVRVPNLKVGFVLLSCAFMYDIFWVFVSKWWFRESVMIVVARG 360
Query 75 FEG-----PAKLMFPVQISPLQ-YSILGLGDVVIPGVLIAMCLRFD 114
P L P P YSI+G GD+++PG+L+ LR+D
Sbjct 361 DRSGEDGIPMLLKIPRMFDPWGGYSIIGFGDIILPGLLVTFALRYD 406
> Hs20514780
Length=564
Score = 58.5 bits (140), Expect = 4e-09, Method: Composition-based stats.
Identities = 43/136 (31%), Positives = 71/136 (52%), Gaps = 26/136 (19%)
Query 6 LVALAICTLWLLTKH-----WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVI 60
L +A+ +W + ++ W L + L IAFC+ + + + +F ++LL LF+YD+
Sbjct 274 LFCVAVSVVWGVFRNEDQWAWVLQDALGIAFCLYMLKTIRLPTFKACTLLLLVLFLYDIF 333
Query 61 WVF--------GTEVMVSVAKAFEG-------PAKLMFP-VQISPLQ-----YSILGLGD 99
+VF G+ +MV VA P L P + SPL +S+LG GD
Sbjct 334 FVFITPFLTKSGSSIMVEVATGPSDSATREKLPMVLKVPRLNSSPLALCDRPFSLLGFGD 393
Query 100 VVIPGVLIAMCLRFDL 115
+++PG+L+A C RFD+
Sbjct 394 ILVPGLLVAYCHRFDI 409
> CE05379
Length=652
Score = 56.6 bits (135), Expect = 1e-08, Method: Composition-based stats.
Identities = 41/157 (26%), Positives = 66/157 (42%), Gaps = 46/157 (29%)
Query 6 LVALAICTLWLLTKH----WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIW 61
++ L+ C W + + + L +++ +A C+ + + + S SIL+ +FVYD
Sbjct 377 IICLSFCVTWFIIRRQPYAFILLDVINMALCMHVLKCLRLPSLKWISILMLCMFVYDAFM 436
Query 62 VFGT--------EVMVSVAKAFEGPAK----------------------LMFPVQISPL- 90
VFGT VM+ VA AK LM +P+
Sbjct 437 VFGTPYMTTNGCSVMLEVATGLSCAAKGKNKGYPVPPIEQESVPEKFPMLMQVAHFNPMN 496
Query 91 -----------QYSILGLGDVVIPGVLIAMCLRFDLF 116
Q++ILGLGD+V+PG L+A C + F
Sbjct 497 ECLDMEIELGFQFTILGLGDIVMPGYLVAHCFTMNGF 533
> At4g33410
Length=311
Score = 55.1 bits (131), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 39/132 (29%), Positives = 61/132 (46%), Gaps = 41/132 (31%)
Query 15 WLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVFGTEVMVSVA-- 72
WL++ HW L+N+L I+ CI ++ V + + + ++LL FG VMV+VA
Sbjct 125 WLISGHWVLNNLLGISICIAFVSHVRLPNIKICAMLL---------RFFGANVMVAVATQ 175
Query 73 ----------------------KAFEGPAKLMFPVQ--------ISPLQYSILGLGDVVI 102
K E P K++FP +S + +LGLGD+ I
Sbjct 176 QASNPVHTVANSLNLPGLQLITKKLELPVKIVFPRNLLGGVVPGVSASDFMMLGLGDMAI 235
Query 103 PGVLIAMCLRFD 114
P +L+A+ L FD
Sbjct 236 PAMLLALVLCFD 247
> CE26879
Length=523
Score = 43.1 bits (100), Expect = 1e-04, Method: Composition-based stats.
Identities = 25/110 (22%), Positives = 57/110 (51%), Gaps = 1/110 (0%)
Query 4 SHLVALAICTLWLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVF 63
S LV+L+ + +W ++ILA A + + S+ A I + G+ ++D+ +++
Sbjct 350 SLLVSLSFGLYHECSGNWISNDILAFASIYVVCSRIQAVSYQTAIIFVIGMSLFDLFFLY 409
Query 64 GTEVMVSVAKAFEGPAKLMFPVQISPLQYSILGLGDVVIPGVLIAMCLRF 113
+++ +V K P ++ P + S+ L D+++PGV + + L++
Sbjct 410 VVDLLSTVVKENRSPLMILVPRDTKGNKQSLAAL-DIMVPGVFLNVVLKY 458
> Hs18587775
Length=684
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 40/125 (32%), Positives = 61/125 (48%), Gaps = 27/125 (21%)
Query 21 WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVF--------GTEVMVSVA 72
W L + L I++C+ + V + + S L L +DV +VF G +M VA
Sbjct 349 WLLQDTLGISYCLFVLHRVRLPTLKNCSSFLLALLAFDVFFVFVTPFFTKTGESIMAQVA 408
Query 73 KAFEGPAK----------LMFP-VQISPL-----QYSILGLGDVVIPGVLIAMCLRFDLF 116
GPA+ L P +++S L +SILG GD+V+PG L+A C RFD+
Sbjct 409 L---GPAESSSHERLPMVLKVPRLRVSALTLCSQPFSILGFGDIVVPGFLVAYCCRFDVQ 465
Query 117 LYLKQ 121
+ +Q
Sbjct 466 VCSRQ 470
> At1g05820
Length=441
Score = 32.0 bits (71), Expect = 0.36, Method: Composition-based stats.
Identities = 11/23 (47%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 92 YSILGLGDVVIPGVLIAMCLRFD 114
Y+++G GD++ PG+LI RFD
Sbjct 335 YNMIGFGDILFPGLLICFIFRFD 357
> CE06465
Length=424
Score = 30.0 bits (66), Expect = 1.3, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 33/67 (49%), Gaps = 12/67 (17%)
Query 9 LAICTLWLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVFGTEVM 68
LAI T +L +W N+++I C+Q +A + + GV+ I Y +W G E++
Sbjct 248 LAIRTPYL---NW---NMISIVCCLQVMACLGIFLVGVSRIF------YGALWAIGIEII 295
Query 69 VSVAKAF 75
+ F
Sbjct 296 FATVALF 302
Lambda K H
0.333 0.143 0.456
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1356426142
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40