bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1670_orf1
Length=104
Score E
Sequences producing significant alignments: (Bits) Value
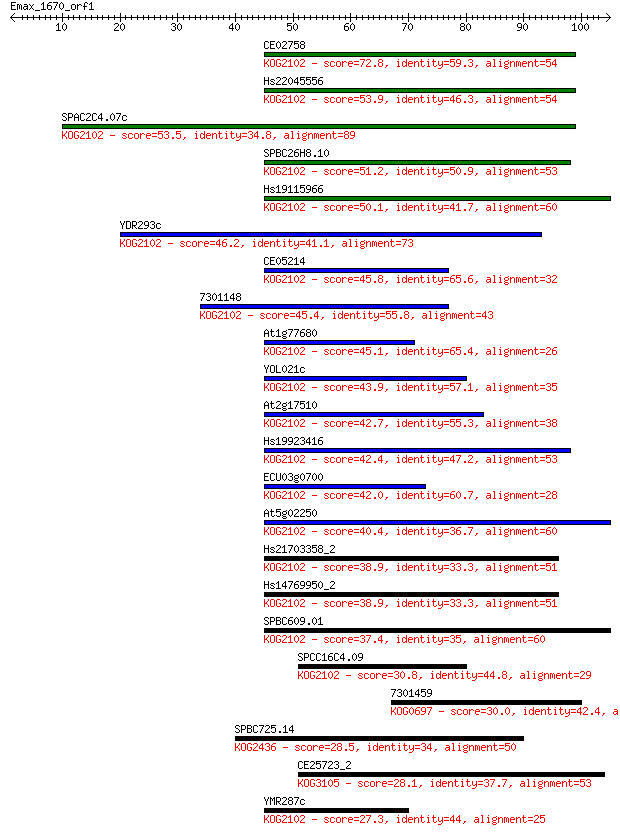
CE02758 72.8 1e-13
Hs22045556 53.9 7e-08
SPAC2C4.07c 53.5 1e-07
SPBC26H8.10 51.2 5e-07
Hs19115966 50.1 1e-06
YDR293c 46.2 2e-05
CE05214 45.8 2e-05
7301148 45.4 3e-05
At1g77680 45.1 3e-05
YOL021c 43.9 8e-05
At2g17510 42.7 2e-04
Hs19923416 42.4 2e-04
ECU03g0700 42.0 3e-04
At5g02250 40.4 7e-04
Hs21703358_2 38.9 0.002
Hs14769950_2 38.9 0.002
SPBC609.01 37.4 0.006
SPCC16C4.09 30.8 0.71
7301459 30.0 1.2
SPBC725.14 28.5 3.3
CE25723_2 28.1 3.8
YMR287c 27.3 6.2
> CE02758
Length=817
Score = 72.8 bits (177), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 32/54 (59%), Positives = 38/54 (70%), Gaps = 0/54 (0%)
Query 45 HFALNFDYYTHSTSPIRRYPDLLVHRTLAALIDNEKHPPFTPEEAEAICNACND 98
HFALN D+YTH TSPIRRYPD++VHR LAA + + PEE + IC CND
Sbjct 623 HFALNVDHYTHFTSPIRRYPDVIVHRQLAAALGYNERSERVPEEIQEICTRCND 676
> Hs22045556
Length=885
Score = 53.9 bits (128), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 25/54 (46%), Positives = 33/54 (61%), Gaps = 0/54 (0%)
Query 45 HFALNFDYYTHSTSPIRRYPDLLVHRTLAALIDNEKHPPFTPEEAEAICNACND 98
H+ALN YTH TSPIRR+ D+LVHR LAA + + P+ + + CND
Sbjct 688 HYALNVPLYTHFTSPIRRFADVLVHRLLAAALGYRERLDMAPDTLQKQADHCND 741
> SPAC2C4.07c
Length=927
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 31/89 (34%), Positives = 50/89 (56%), Gaps = 4/89 (4%)
Query 10 QLIGVYTEVLLRTFLSLAEYKTIKQIEEFINHKKIHFALNFDYYTHSTSPIRRYPDLLVH 69
+L+ ++ + +R+ L+ AEY E + H+AL+F++YTH TSPIRRYPD++VH
Sbjct 722 ELVELFENMAVRS-LNRAEYFCTGDFGEKTDWH--HYALSFNHYTHFTSPIRRYPDIIVH 778
Query 70 RTLAALIDNEKHPPFTPEEAEAICNACND 98
R L + N P + + CN+
Sbjct 779 RLLERSLKNTS-PGIDKKNCSLVAAHCNE 806
> SPBC26H8.10
Length=970
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 27/55 (49%), Positives = 32/55 (58%), Gaps = 2/55 (3%)
Query 45 HFALNFDYYTHSTSPIRRYPDLLVHRTLAALIDNEKHPPFTPEEAE--AICNACN 97
H+ L YTH TSPIRRY D+L HR LAA ID E P +++ ICN N
Sbjct 796 HYGLASPIYTHFTSPIRRYADVLAHRQLAAAIDYETINPSLSDKSRLIEICNGIN 850
> Hs19115966
Length=971
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 37/65 (56%), Gaps = 5/65 (7%)
Query 45 HFALNFDYYTHSTSPIRRYPDLLVHRTLAALIDNEKHPP-----FTPEEAEAICNACNDV 99
H+ L D YTH TSPIRRY D++VHR L A I +K F+ ++ E +C N+
Sbjct 701 HYGLALDKYTHFTSPIRRYSDIVVHRLLMAAISKDKKMEIKGNLFSNKDLEELCRHINNR 760
Query 100 GKSMR 104
++ +
Sbjct 761 NQAAQ 765
> YDR293c
Length=1250
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/78 (38%), Positives = 45/78 (57%), Gaps = 9/78 (11%)
Query 20 LRTFLSLAEYKTIKQIEEFINHKKI-----HFALNFDYYTHSTSPIRRYPDLLVHRTLAA 74
+R + + +KT+ + FI K H+ALN YTH T+P+RRY D +VHR L A
Sbjct 953 VRVGIEILLFKTMPRARYFIAGKVDPDQYGHYALNLPIYTHFTAPMRRYADHVVHRQLKA 1012
Query 75 LIDNEKHPPFTPEEAEAI 92
+I + P+T E+ EA+
Sbjct 1013 VIHDT---PYT-EDMEAL 1026
> CE05214
Length=1029
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 21/32 (65%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 45 HFALNFDYYTHSTSPIRRYPDLLVHRTLAALI 76
HF L YTH TSPIRRY D++VHR LAA I
Sbjct 846 HFGLACAIYTHFTSPIRRYADVIVHRLLAAAI 877
> 7301148
Length=982
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 24/43 (55%), Positives = 27/43 (62%), Gaps = 5/43 (11%)
Query 34 QIEEFINHKKIHFALNFDYYTHSTSPIRRYPDLLVHRTLAALI 76
Q EEF H+ L YTH TSPIRRY D++VHR LAA I
Sbjct 767 QKEEFF-----HYGLAAPIYTHFTSPIRRYSDIMVHRLLAASI 804
> At1g77680
Length=1055
Score = 45.1 bits (105), Expect = 3e-05, Method: Composition-based stats.
Identities = 17/26 (65%), Positives = 21/26 (80%), Gaps = 0/26 (0%)
Query 45 HFALNFDYYTHSTSPIRRYPDLLVHR 70
H+AL YTH TSP+RRYPD++VHR
Sbjct 792 HYALAVPLYTHFTSPLRRYPDIVVHR 817
> YOL021c
Length=1001
Score = 43.9 bits (102), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 20/35 (57%), Positives = 23/35 (65%), Gaps = 0/35 (0%)
Query 45 HFALNFDYYTHSTSPIRRYPDLLVHRTLAALIDNE 79
H+ L D YTH TSPIRRY D++ HR LA I E
Sbjct 831 HYGLAVDIYTHFTSPIRRYCDVVAHRQLAGAIGYE 865
> At2g17510
Length=955
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/38 (55%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 45 HFALNFDYYTHSTSPIRRYPDLLVHRTLAALIDNEKHP 82
H+ L YTH TSPIRRY D+ VHR LAA + K P
Sbjct 789 HYGLAAPLYTHFTSPIRRYADVFVHRLLAASLGIYKLP 826
> Hs19923416
Length=958
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/55 (45%), Positives = 31/55 (56%), Gaps = 2/55 (3%)
Query 45 HFALNFDYYTHSTSPIRRYPDLLVHRTLAALIDNE-KHPPFTPEEAEA-ICNACN 97
H+ L YTH TSPIRRY D++VHR LA I + +P T + A IC N
Sbjct 764 HYGLASPIYTHFTSPIRRYADVIVHRLLAVAIGADCTYPELTDKHKLADICKNLN 818
> ECU03g0700
Length=835
Score = 42.0 bits (97), Expect = 3e-04, Method: Composition-based stats.
Identities = 17/28 (60%), Positives = 20/28 (71%), Gaps = 0/28 (0%)
Query 45 HFALNFDYYTHSTSPIRRYPDLLVHRTL 72
H+ L YTH TSPIRRY D++VHR L
Sbjct 667 HYGLATPIYTHFTSPIRRYADIIVHRIL 694
> At5g02250
Length=782
Score = 40.4 bits (93), Expect = 7e-04, Method: Composition-based stats.
Identities = 22/60 (36%), Positives = 28/60 (46%), Gaps = 0/60 (0%)
Query 45 HFALNFDYYTHSTSPIRRYPDLLVHRTLAALIDNEKHPPFTPEEAEAICNACNDVGKSMR 104
H L Y TSPIRRY DL H + A + + PF+ E E I + N K +R
Sbjct 641 HGVLGIPGYVQFTSPIRRYMDLTAHYQIKAFLRGGDNFPFSAGELEGIAASVNMQSKVVR 700
> Hs21703358_2
Length=952
Score = 38.9 bits (89), Expect = 0.002, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 30/51 (58%), Gaps = 1/51 (1%)
Query 45 HFALNFDYYTHSTSPIRRYPDLLVHRTLAALIDNEKHPPFTPEEAEAICNA 95
H++L D+YT +TSPIRRY D+++ R + L ++ + + +C A
Sbjct 461 HYSLQVDWYTWATSPIRRYLDVVLQRQI-LLALGHGGSAYSARDIDGLCQA 510
> Hs14769950_2
Length=968
Score = 38.9 bits (89), Expect = 0.002, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 30/51 (58%), Gaps = 1/51 (1%)
Query 45 HFALNFDYYTHSTSPIRRYPDLLVHRTLAALIDNEKHPPFTPEEAEAICNA 95
H++L D+YT +TSPIRRY D+++ R + L ++ + + +C A
Sbjct 477 HYSLQVDWYTWATSPIRRYLDVVLQRQI-LLALGHGGSAYSARDIDGLCQA 526
> SPBC609.01
Length=1157
Score = 37.4 bits (85), Expect = 0.006, Method: Composition-based stats.
Identities = 21/62 (33%), Positives = 28/62 (45%), Gaps = 4/62 (6%)
Query 45 HFALNFDYYTHSTSPIRRYPDLLVHRTLAALIDNEKHPPFTPEEAE--AICNACNDVGKS 102
HF + YYTH P RRY D+ V R L D P F+ + +I +CN +
Sbjct 891 HFVFSCPYYTHFCHPTRRYIDICVQRQLREAFDGR--PDFSKDYRSLLSITQSCNTLSNF 948
Query 103 MR 104
R
Sbjct 949 YR 950
> SPCC16C4.09
Length=1066
Score = 30.8 bits (68), Expect = 0.71, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 51 DYYTHSTSPIRRYPDLLVHRTLAALIDNE 79
D TH T+P+ RY D++VH L L+ E
Sbjct 908 DDLTHFTAPLERYGDIVVHYQLQLLLRGE 936
> 7301459
Length=367
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 67 LVHRTLAALIDNEKHPPFTPEEAEAICNACNDV 99
++ R AA+I H PF+P+E E I NA V
Sbjct 139 VISRNGAAVISTIDHKPFSPKEQERIQNAGGSV 171
> SPBC725.14
Length=500
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 23/50 (46%), Gaps = 0/50 (0%)
Query 40 NHKKIHFALNFDYYTHSTSPIRRYPDLLVHRTLAALIDNEKHPPFTPEEA 89
+H I+ A FD + P R +LV R L L D+ TPE+A
Sbjct 238 SHVLINLAQEFDELAKTLPPYHRKNLILVRRCLKMLPDDASALITTPEDA 287
> CE25723_2
Length=271
Score = 28.1 bits (61), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 27/56 (48%), Gaps = 3/56 (5%)
Query 51 DYYTHS--TSPIRR-YPDLLVHRTLAALIDNEKHPPFTPEEAEAICNACNDVGKSM 103
D Y +S + IRR Y D +VH + N PP + E I NA ND+ K +
Sbjct 142 DVYWNSPFKASIRRSYEDWMVHGQKSFTKSNNMRPPSMLQYLEWIENAWNDLPKDL 197
> YMR287c
Length=969
Score = 27.3 bits (59), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 11/25 (44%), Positives = 15/25 (60%), Gaps = 0/25 (0%)
Query 45 HFALNFDYYTHSTSPIRRYPDLLVH 69
H + Y TSP+RR+PDL+ H
Sbjct 821 HEMIGAKQYLTVTSPLRRFPDLINH 845
Lambda K H
0.323 0.138 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174332180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40