bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1655_orf1
Length=124
Score E
Sequences producing significant alignments: (Bits) Value
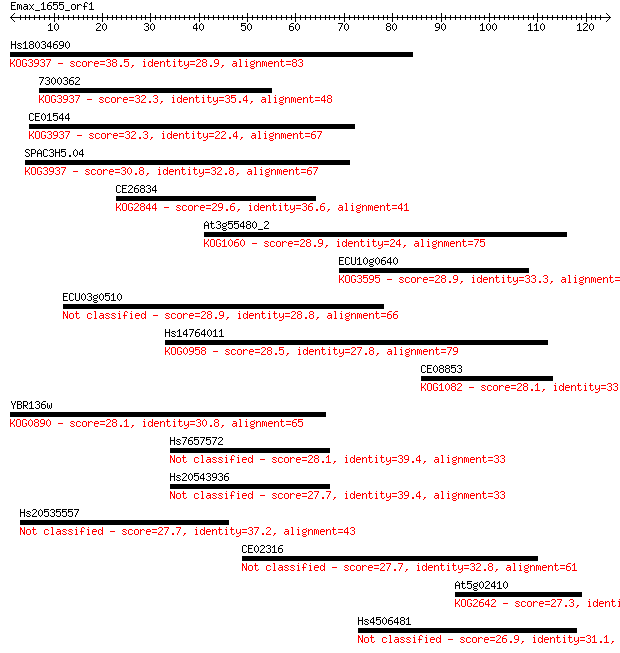
Hs18034690 38.5 0.003
7300362 32.3 0.20
CE01544 32.3 0.22
SPAC3H5.04 30.8 0.59
CE26834 29.6 1.5
At3g55480_2 28.9 2.3
ECU10g0640 28.9 2.4
ECU03g0510 28.9 2.7
Hs14764011 28.5 3.0
CE08853 28.1 4.2
YBR136w 28.1 4.4
Hs7657572 28.1 4.5
Hs20543936 27.7 4.7
Hs20535557 27.7 5.1
CE02316 27.7 5.7
At5g02410 27.3 7.4
Hs4506481 26.9 9.6
> Hs18034690
Length=384
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 24/85 (28%), Positives = 41/85 (48%), Gaps = 2/85 (2%)
Query 1 LAHHFPSLLHWRELIQLLSNAGRAVYLLPGLYGKFLDTFYAQLSQSPDDIILGPLEEGSF 60
L + + + HW+ L+ LL + A+ LY + Y QL + P D + + + +F
Sbjct 264 LGNVYEAFEHWKRLLNLLCRSEAAMMKHHTLYINLISILYHQLGEIPADFFVDIVSQDNF 323
Query 61 LSSSCASLLEN-CSY-IDRQLAKKA 83
L+S+ + CS +D L KKA
Sbjct 324 LTSTLQVFFSSACSIAVDATLRKKA 348
> 7300362
Length=379
Score = 32.3 bits (72), Expect = 0.20, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 29/49 (59%), Gaps = 1/49 (2%)
Query 7 SLLHWRELIQLLSNAGRAVYLLPGLYGKFLDTFYAQLSQSPDDIIL-GP 54
SL HWR+L+ LL+++ AV Y K+ + QL P+++++ GP
Sbjct 272 SLAHWRKLLGLLAHSQTAVTKHKLTYMKYSEVLAHQLPHLPEELMVPGP 320
> CE01544
Length=357
Score = 32.3 bits (72), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 15/67 (22%), Positives = 31/67 (46%), Gaps = 0/67 (0%)
Query 5 FPSLLHWRELIQLLSNAGRAVYLLPGLYGKFLDTFYAQLSQSPDDIILGPLEEGSFLSSS 64
F W+ +I L+S ++ L+ F+ + QL + P D + + +FL+++
Sbjct 258 FEGFEQWKRIIHLMSCCPNSLGSEKELFMSFIRVLFFQLKECPTDFFVDIVSRDNFLTTT 317
Query 65 CASLLEN 71
+ L N
Sbjct 318 LSMLFAN 324
> SPAC3H5.04
Length=346
Score = 30.8 bits (68), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 31/68 (45%), Gaps = 1/68 (1%)
Query 4 HFPSLLHWRELIQLLSNAGRAVYLLPGLYGKFLDTFYAQLSQ-SPDDIILGPLEEGSFLS 62
H+ +L HW+ ++ LL + P Y FL+ F QLS S D+ + E L
Sbjct 210 HYGALEHWKNMLSLLLQSYELAETEPEFYASFLELFKLQLSSLSESDLETSAIFEKGVLL 269
Query 63 SSCASLLE 70
S SL E
Sbjct 270 SCLDSLSE 277
> CE26834
Length=725
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 23 RAVYLLPGLYGKFLDTFYAQLSQSPDDI-ILGPLEEGSFLSS 63
RA L+PGL G +D A S +PD ++GP ++ ++S+
Sbjct 332 RACDLIPGLQGSKVDARAAVFSMTPDGYPLVGPYDKNYWMST 373
> At3g55480_2
Length=1113
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 18/77 (23%), Positives = 34/77 (44%), Gaps = 2/77 (2%)
Query 41 AQLSQSPDDIILGPLEEGSFLSSSCASLLENCSYIDRQLAKKALIQQITNNI--QEKKQK 98
A L P+D+ + PL + F S C +L + I + Q+ N+ Q + K
Sbjct 3 AHLYDDPEDVNIAPLLDSKFESEKCEALKRLLALIAQGFDVSNFFPQVVKNVASQSSEVK 62
Query 99 KIQNIHQLSNKKGKGNK 115
K+ ++ L + + N+
Sbjct 63 KLVYLYLLQYAEKRPNE 79
> ECU10g0640
Length=3151
Score = 28.9 bits (63), Expect = 2.4, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 69 LENCSYIDRQLAKKALIQQITNNIQEKKQKKIQNIHQLS 107
LEN +DRQ AKK + ++ N++ +K K ++ +S
Sbjct 2879 LENERNVDRQYAKKREVYEVLNSLFSRKSKSFFEVYGVS 2917
> ECU03g0510
Length=1243
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 19/68 (27%), Positives = 33/68 (48%), Gaps = 2/68 (2%)
Query 12 RELIQLLSNAGRAVYLLPGLYGKFLDTFYAQ--LSQSPDDIILGPLEEGSFLSSSCASLL 69
R+++ + RA +P F+ TF L+ S + I+G + EGS+ +S +L
Sbjct 51 RDVVDVFVQGLRAKRQVPAFVIDFIYTFLVNDLLTSSDIEEIIGVISEGSWCEASKMKIL 110
Query 70 ENCSYIDR 77
+ YI R
Sbjct 111 QMSYYIMR 118
> Hs14764011
Length=1096
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 22/79 (27%), Positives = 34/79 (43%), Gaps = 12/79 (15%)
Query 33 GKFLDTFYAQLSQSPDDIILGPLEEGSFLSSSCASLLENCSYIDRQLAKKALIQQITNNI 92
G + D Y + S D + LGP EG L + D L K I +T++I
Sbjct 955 GSYSDNLYPESITSRDCVQLGPPSEGE---------LVELRWTDGNLYKAKFISSVTSHI 1005
Query 93 QEKKQKKIQNIHQLSNKKG 111
Q + ++ QL+ K+G
Sbjct 1006 Y---QVEFEDGSQLTVKRG 1021
> CE08853
Length=737
Score = 28.1 bits (61), Expect = 4.2, Method: Composition-based stats.
Identities = 9/27 (33%), Positives = 21/27 (77%), Gaps = 0/27 (0%)
Query 86 QQITNNIQEKKQKKIQNIHQLSNKKGK 112
+ I+NN+ EK+++ +++ H+L +K+ K
Sbjct 89 RAISNNLTEKRRRSVESFHRLHSKERK 115
> YBR136w
Length=2368
Score = 28.1 bits (61), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 31/65 (47%), Gaps = 5/65 (7%)
Query 1 LAHHFPSLLHWRELIQLLSNAGRAVYLLPGLYGKFLDTFYAQLSQSPDDIILGPLEEGSF 60
LA +PS + W + SN+ + V L GK + Y Q SQ+P D++ L+
Sbjct 1926 LAVEYPSHILWYITALVNSNSSKRV-----LRGKHILEKYRQHSQNPHDLVSSALDLTKA 1980
Query 61 LSSSC 65
L+ C
Sbjct 1981 LTRVC 1985
> Hs7657572
Length=431
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 18/33 (54%), Gaps = 0/33 (0%)
Query 34 KFLDTFYAQLSQSPDDIILGPLEEGSFLSSSCA 66
K L F L+ PD +ILG LE G + +C+
Sbjct 144 KQLSVFVTALTHRPDILILGTLESGHSRNLTCS 176
> Hs20543936
Length=499
Score = 27.7 bits (60), Expect = 4.7, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 18/33 (54%), Gaps = 0/33 (0%)
Query 34 KFLDTFYAQLSQSPDDIILGPLEEGSFLSSSCA 66
K L F L+ PD +ILG LE G + +C+
Sbjct 144 KQLSVFVTALTHRPDILILGTLESGHSRNLTCS 176
> Hs20535557
Length=482
Score = 27.7 bits (60), Expect = 5.1, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 23/43 (53%), Gaps = 1/43 (2%)
Query 3 HHFPSLLHWRELIQLLSNAGRAVYLLPGLYGKFLDTFYAQLSQ 45
H+F L W ELI L S G + L+PG +G + ++ SQ
Sbjct 421 HNF-YLFPWGELIALPSIVGPPLLLIPGFHGLAVLSYSCSRSQ 462
> CE02316
Length=649
Score = 27.7 bits (60), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 33/65 (50%), Gaps = 4/65 (6%)
Query 49 DIILGPLEEGSFLSSSCASLLENCSYIDRQLAK----KALIQQITNNIQEKKQKKIQNIH 104
+I + PL E +F +S + +N ID L K K + I N ++++ IQN+H
Sbjct 205 EIPIRPLAEVTFSGTSDKFITKNRPIIDSNLNKVSDEKEEVDDIINLSGDQEEVSIQNMH 264
Query 105 QLSNK 109
Q + K
Sbjct 265 QPTKK 269
> At5g02410
Length=469
Score = 27.3 bits (59), Expect = 7.4, Method: Composition-based stats.
Identities = 12/26 (46%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 93 QEKKQKKIQNIHQLSNKKGKGNKNNI 118
Q+ KQ+ Q +HQ SNKKG ++N+
Sbjct 206 QKGKQEVNQELHQSSNKKGATLRSNL 231
> Hs4506481
Length=1224
Score = 26.9 bits (58), Expect = 9.6, Method: Composition-based stats.
Identities = 14/45 (31%), Positives = 25/45 (55%), Gaps = 1/45 (2%)
Query 73 SYIDRQLAKKALIQQITNNIQEKKQKKIQNIHQLSNKKGKGNKNN 117
+Y+D Q+ K + +T + + KQ++ QL+NKK +NN
Sbjct 141 AYVDEQMTMKTKQRMLTEDWELFKQRRFIE-EQLTNKKAVTGENN 184
Lambda K H
0.318 0.135 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187579072
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40