bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1629_orf1
Length=89
Score E
Sequences producing significant alignments: (Bits) Value
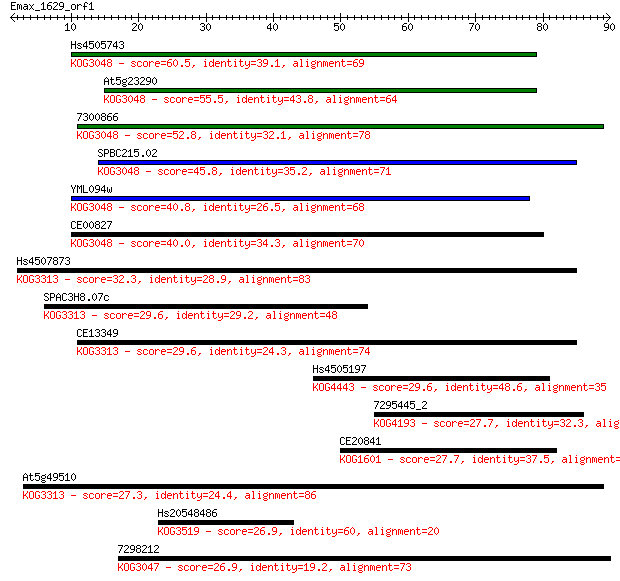
Hs4505743 60.5 7e-10
At5g23290 55.5 2e-08
7300866 52.8 1e-07
SPBC215.02 45.8 2e-05
YML094w 40.8 6e-04
CE00827 40.0 0.001
Hs4507873 32.3 0.24
SPAC3H8.07c 29.6 1.3
CE13349 29.6 1.4
Hs4505197 29.6 1.6
7295445_2 27.7 5.5
CE20841 27.7 5.6
At5g49510 27.3 7.2
Hs20548486 26.9 8.2
7298212 26.9 8.5
> Hs4505743
Length=167
Score = 60.5 bits (145), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 27/69 (39%), Positives = 45/69 (65%), Gaps = 0/69 (0%)
Query 10 GETAEVLVPLTGALYAKGRLQCDDKVLVDIGAGYMLQKTCKDAKKDGERTLNLVSEQQGK 69
E E+LVPLT ++Y G+L + VL+D+G GY ++KT +DAK +R ++ +++Q K
Sbjct 70 NEGKELLVPLTSSMYVPGKLHDVEHVLIDVGTGYYVEKTAEDAKDFFKRKIDFLTKQMEK 129
Query 70 LEKMLSEKQ 78
++ L EK
Sbjct 130 IQPALQEKH 138
> At5g23290
Length=151
Score = 55.5 bits (132), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 28/64 (43%), Positives = 41/64 (64%), Gaps = 0/64 (0%)
Query 15 VLVPLTGALYAKGRLQCDDKVLVDIGAGYMLQKTCKDAKKDGERTLNLVSEQQGKLEKML 74
+LVPLT +LY G L DKVLVDIG GY ++KT D K +R +NL+ +L ++
Sbjct 65 MLVPLTASLYVPGTLDEADKVLVDIGTGYFIEKTMDDGKDYCQRKINLLKSNFDQLFEVA 124
Query 75 SEKQ 78
++K+
Sbjct 125 AKKK 128
> 7300866
Length=168
Score = 52.8 bits (125), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/78 (32%), Positives = 48/78 (61%), Gaps = 0/78 (0%)
Query 11 ETAEVLVPLTGALYAKGRLQCDDKVLVDIGAGYMLQKTCKDAKKDGERTLNLVSEQQGKL 70
E ++LVPLT ++Y GR++ ++ ++DIG GY ++K + +K +R + V EQ K+
Sbjct 67 ENRQILVPLTSSMYVPGRVKDLNRFVIDIGTGYYIEKDLEGSKDYFKRRVEYVQEQIEKI 126
Query 71 EKMLSEKQKQLEVLVATL 88
EK+ +K + +++ L
Sbjct 127 EKIHLQKTRFYNSVMSVL 144
> SPBC215.02
Length=154
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 40/72 (55%), Gaps = 1/72 (1%)
Query 14 EVLVPLTGALYAKGRLQC-DDKVLVDIGAGYMLQKTCKDAKKDGERTLNLVSEQQGKLEK 72
EVLVPLT +LY G+L + K+LVDIG GY ++K+ +A + +R ++ L
Sbjct 65 EVLVPLTSSLYVPGKLNLGNSKLLVDIGTGYYVEKSAGEATEYYKRKCEYLASSIENLNN 124
Query 73 MLSEKQKQLEVL 84
+ K Q+ +
Sbjct 125 AIDAKSVQIRAV 136
> YML094w
Length=163
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 43/68 (63%), Gaps = 0/68 (0%)
Query 10 GETAEVLVPLTGALYAKGRLQCDDKVLVDIGAGYMLQKTCKDAKKDGERTLNLVSEQQGK 69
E ++LVP + +LY G++ + K +VDIG GY ++K+ + A ++ ++ ++++ +
Sbjct 59 NEGQKLLVPASASLYIPGKIVDNKKFMVDIGTGYYVEKSAEAAIAFYQKKVDKLNKESVQ 118
Query 70 LEKMLSEK 77
++ ++ EK
Sbjct 119 IQDIIKEK 126
> CE00827
Length=152
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 40/70 (57%), Gaps = 2/70 (2%)
Query 10 GETAEVLVPLTGALYAKGRLQCDDKVLVDIGAGYMLQKTCKDAKKDGERTLNLVSEQQGK 69
G TA L+PL+ +LY + L K LV+IG GY ++ + AK +R +++Q
Sbjct 62 GHTA--LIPLSESLYIRAELSDPSKHLVEIGTGYFVELDREKAKAIFDRKKEHITKQVET 119
Query 70 LEKMLSEKQK 79
+E +L EK++
Sbjct 120 VEGILKEKRR 129
> Hs4507873
Length=197
Score = 32.3 bits (72), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 24/83 (28%), Positives = 36/83 (43%), Gaps = 14/83 (16%)
Query 2 HIQRQKQKGETAEVLVPLTGALYAKGRLQCDDKVLVDIGAGYMLQKTCKDAKKDGERTLN 61
++Q++K+ + E L LY K + DKV + +GA ML+
Sbjct 90 YMQKKKESTNSMETRFLLADNLYCKASVPPTDKVCLWLGANVMLEYD------------- 136
Query 62 LVSEQQGKLEKMLSEKQKQLEVL 84
+ E Q LEK LS K L+ L
Sbjct 137 -IDEAQALLEKNLSTATKNLDSL 158
> SPAC3H8.07c
Length=169
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 28/48 (58%), Gaps = 0/48 (0%)
Query 6 QKQKGETAEVLVPLTGALYAKGRLQCDDKVLVDIGAGYMLQKTCKDAK 53
++++G++ V L L AK ++ D V + +GA ML+ T ++A+
Sbjct 71 KERQGDSFTVTYELNDTLNAKAEVEAKDNVYLWLGANVMLEYTVEEAE 118
> CE13349
Length=185
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 18/74 (24%), Positives = 39/74 (52%), Gaps = 7/74 (9%)
Query 11 ETAEVLVPLTGALYAKGRLQCDDKVLVDIGAGYMLQKTCKDAKKDGERTLNLVSEQQGKL 70
E+ E L+ +Y K +Q +KV + +GA M++ ++A+K L+ + +G +
Sbjct 86 ESFETTFLLSDDVYTKATVQKPEKVSIWLGANVMVEYDLENARK-------LLDKNRGSV 138
Query 71 EKMLSEKQKQLEVL 84
+K++ E +L +
Sbjct 139 QKVVDELTNELSYI 152
> Hs4505197
Length=5262
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 17/40 (42%), Positives = 25/40 (62%), Gaps = 6/40 (15%)
Query 46 QKTCKDAKK-----DGERTLNLVSEQQGKLEKMLSEKQKQ 80
Q+T K A + D E+ L LV+EQQ K++K L + +KQ
Sbjct 3271 QRTAKKAGREFPEADAEK-LKLVTEQQSKIQKQLDQVRKQ 3309
> 7295445_2
Length=282
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 10/31 (32%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 55 DGERTLNLVSEQQGKLEKMLSEKQKQLEVLV 85
D +RTL ++ +G+++K+ S+KQ + L+
Sbjct 193 DSKRTLRKIARNEGRIQKLNSDKQNYTQFLL 223
> CE20841
Length=403
Score = 27.7 bits (60), Expect = 5.6, Method: Composition-based stats.
Identities = 12/32 (37%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 50 KDAKKDGERTLNLVSEQQGKLEKMLSEKQKQL 81
KD ++D + ++ + E G+LE ++SEK+K L
Sbjct 43 KDIRQDVSKMMSKIDEVCGRLEALISEKEKVL 74
> At5g49510
Length=195
Score = 27.3 bits (59), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 21/86 (24%), Positives = 39/86 (45%), Gaps = 14/86 (16%)
Query 3 IQRQKQKGETAEVLVPLTGALYAKGRLQCDDKVLVDIGAGYMLQKTCKDAKKDGERTLNL 62
++ +K GE ++ +Y++ ++ D V + +GA ML+ +C++A
Sbjct 84 LEAKKGTGEALLADFEVSEGIYSRACIEDTDSVCLWLGANVMLEYSCEEAS--------- 134
Query 63 VSEQQGKLEKMLSEKQKQLEVLVATL 88
L+ L + LEVLVA L
Sbjct 135 -----ALLKNNLENAKASLEVLVADL 155
> Hs20548486
Length=652
Score = 26.9 bits (58), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 12/20 (60%), Positives = 14/20 (70%), Gaps = 0/20 (0%)
Query 23 LYAKGRLQCDDKVLVDIGAG 42
LY KGRL D+ LVD+G G
Sbjct 496 LYYKGRLDMDEMELVDLGDG 515
> 7298212
Length=150
Score = 26.9 bits (58), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 14/73 (19%), Positives = 39/73 (53%), Gaps = 0/73 (0%)
Query 17 VPLTGALYAKGRLQCDDKVLVDIGAGYMLQKTCKDAKKDGERTLNLVSEQQGKLEKMLSE 76
V + ++ + R++ D +LVD+G L + DA++ + + ++++Q L +
Sbjct 66 VNIGSNVFMQARVRKMDSILVDVGKNVFLDMSIPDAERFCDTRVKILTKQSDVLRDESVK 125
Query 77 KQKQLEVLVATLN 89
K+ Q+++ + ++
Sbjct 126 KRAQIKMALIAIS 138
Lambda K H
0.312 0.130 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1181033294
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40