bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
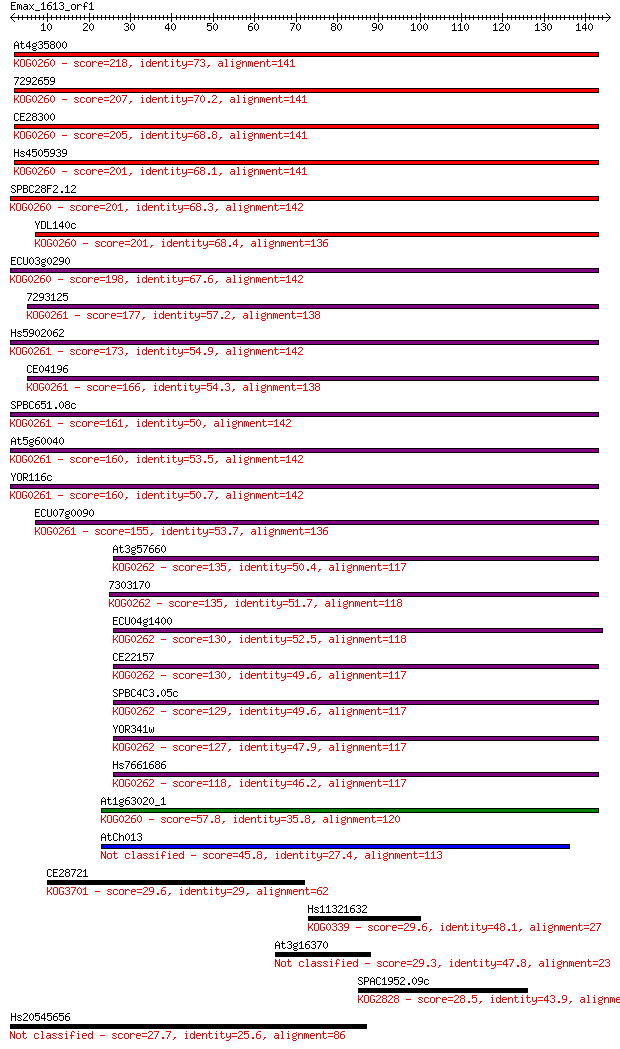
Query= Emax_1613_orf1
Length=145
Score E
Sequences producing significant alignments: (Bits) Value
At4g35800 218 5e-57
7292659 207 4e-54
CE28300 205 2e-53
Hs4505939 201 4e-52
SPBC28F2.12 201 5e-52
YDL140c 201 6e-52
ECU03g0290 198 2e-51
7293125 177 4e-45
Hs5902062 173 1e-43
CE04196 166 1e-41
SPBC651.08c 161 4e-40
At5g60040 160 1e-39
YOR116c 160 1e-39
ECU07g0090 155 4e-38
At3g57660 135 3e-32
7303170 135 3e-32
ECU04g1400 130 9e-31
CE22157 130 1e-30
SPBC4C3.05c 129 1e-30
YOR341w 127 6e-30
Hs7661686 118 3e-27
At1g63020_1 57.8 8e-09
AtCh013 45.8 3e-05
CE28721 29.6 1.9
Hs11321632 29.6 2.0
At3g16370 29.3 2.6
SPAC1952.09c 28.5 4.2
Hs20545656 27.7 6.9
> At4g35800
Length=1840
Score = 218 bits (554), Expect = 5e-57, Method: Compositional matrix adjust.
Identities = 103/141 (73%), Positives = 117/141 (82%), Gaps = 0/141 (0%)
Query 2 RVNKELNAAREKSAKVASEQLDESNNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRIP 61
RVN+ LN AR+ + A + L E+NN+ AMV AGSKGS INISQ+ ACVGQQNVEGKRIP
Sbjct 737 RVNQVLNKARDDAGSSAQKSLAETNNLKAMVTAGSKGSFINISQMTACVGQQNVEGKRIP 796
Query 62 SGFNNRSLPHFSKFDYPPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGY 121
GF+ R+LPHF+K DY PESRGFVE+SYL GL PQE FFHAMGGREG+IDTA KTSETGY
Sbjct 797 FGFDGRTLPHFTKDDYGPESRGFVENSYLRGLTPQEFFFHAMGGREGLIDTAVKTSETGY 856
Query 122 IQRRLMKAMEDVKVSYDRTVR 142
IQRRL+KAMED+ V YD TVR
Sbjct 857 IQRRLVKAMEDIMVKYDGTVR 877
> 7292659
Length=1887
Score = 207 bits (528), Expect = 4e-54, Method: Compositional matrix adjust.
Identities = 99/141 (70%), Positives = 116/141 (82%), Gaps = 0/141 (0%)
Query 2 RVNKELNAAREKSAKVASEQLDESNNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRIP 61
+VN+ LN AR+K+ A + L E NN+ AMV++GSKGS INISQ++ACVGQQNVEGKRIP
Sbjct 732 KVNRILNDARDKTGGSAKKSLTEYNNLKAMVVSGSKGSNINISQVIACVGQQNVEGKRIP 791
Query 62 SGFNNRSLPHFSKFDYPPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGY 121
GF R+LPHF K DY PESRGFVE+SYL GL P E +FHAMGGREG+IDTA KT+ETGY
Sbjct 792 YGFRKRTLPHFIKDDYGPESRGFVENSYLAGLTPSEFYFHAMGGREGLIDTAVKTAETGY 851
Query 122 IQRRLMKAMEDVKVSYDRTVR 142
IQRRL+KAME V V+YD TVR
Sbjct 852 IQRRLIKAMESVMVNYDGTVR 872
> CE28300
Length=1852
Score = 205 bits (522), Expect = 2e-53, Method: Compositional matrix adjust.
Identities = 97/141 (68%), Positives = 116/141 (82%), Gaps = 0/141 (0%)
Query 2 RVNKELNAAREKSAKVASEQLDESNNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRIP 61
+VN+ LN AR+++ A + L E NN +MV++GSKGS INISQ++ACVGQQNVEGKRIP
Sbjct 730 KVNQILNDARDRTGSSAQKSLSEFNNFKSMVVSGSKGSKINISQVIACVGQQNVEGKRIP 789
Query 62 SGFNNRSLPHFSKFDYPPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGY 121
GF +R+LPHF K DY PES+GFVE+SYL GL P E FFHAMGGREG+IDTA KT+ETGY
Sbjct 790 FGFRHRTLPHFIKDDYGPESKGFVENSYLAGLTPSEFFFHAMGGREGLIDTAVKTAETGY 849
Query 122 IQRRLMKAMEDVKVSYDRTVR 142
IQRRL+KAME V V+YD TVR
Sbjct 850 IQRRLIKAMESVMVNYDGTVR 870
> Hs4505939
Length=1970
Score = 201 bits (511), Expect = 4e-52, Method: Compositional matrix adjust.
Identities = 96/141 (68%), Positives = 114/141 (80%), Gaps = 0/141 (0%)
Query 2 RVNKELNAAREKSAKVASEQLDESNNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRIP 61
+VN+ LN AR+K+ A + L E NN +MV++G+KGS INISQ++A VGQQNVEGKRIP
Sbjct 740 QVNRILNDARDKTGSSAQKSLSEYNNFKSMVVSGAKGSKINISQVIAVVGQQNVEGKRIP 799
Query 62 SGFNNRSLPHFSKFDYPPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGY 121
GF +R+LPHF K DY PESRGFVE+SYL GL P E FFHAMGGREG+IDTA KT+ETGY
Sbjct 800 FGFKHRTLPHFIKDDYGPESRGFVENSYLAGLTPTEFFFHAMGGREGLIDTAVKTAETGY 859
Query 122 IQRRLMKAMEDVKVSYDRTVR 142
IQRRL+K+ME V V YD TVR
Sbjct 860 IQRRLIKSMESVMVKYDATVR 880
> SPBC28F2.12
Length=1752
Score = 201 bits (510), Expect = 5e-52, Method: Compositional matrix adjust.
Identities = 97/142 (68%), Positives = 112/142 (78%), Gaps = 0/142 (0%)
Query 1 ARVNKELNAAREKSAKVASEQLDESNNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRI 60
A+V++ LN AR+ + + A L +SNN+ MV AGSKGS INISQ+ ACVGQQ VEGKRI
Sbjct 722 AKVSRILNQARDNAGRSAEHSLKDSNNVKQMVAAGSKGSFINISQMSACVGQQIVEGKRI 781
Query 61 PSGFNNRSLPHFSKFDYPPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETG 120
P GF R+LPHF K D PESRGF+E+SYL GL PQE FFHAM GREG+IDTA KT+ETG
Sbjct 782 PFGFKYRTLPHFPKDDDSPESRGFIENSYLRGLTPQEFFFHAMAGREGLIDTAVKTAETG 841
Query 121 YIQRRLMKAMEDVKVSYDRTVR 142
YIQRRL+KAMEDV V YD TVR
Sbjct 842 YIQRRLVKAMEDVMVRYDGTVR 863
> YDL140c
Length=1733
Score = 201 bits (510), Expect = 6e-52, Method: Compositional matrix adjust.
Identities = 93/136 (68%), Positives = 112/136 (82%), Gaps = 0/136 (0%)
Query 7 LNAAREKSAKVASEQLDESNNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRIPSGFNN 66
LN AR+K+ ++A L + NN+ MV+AGSKGS INI+Q+ ACVGQQ+VEGKRI GF +
Sbjct 722 LNEARDKAGRLAEVNLKDLNNVKQMVMAGSKGSFINIAQMSACVGQQSVEGKRIAFGFVD 781
Query 67 RSLPHFSKFDYPPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRL 126
R+LPHFSK DY PES+GFVE+SYL GL PQE FFHAMGGREG+IDTA KT+ETGYIQRRL
Sbjct 782 RTLPHFSKDDYSPESKGFVENSYLRGLTPQEFFFHAMGGREGLIDTAVKTAETGYIQRRL 841
Query 127 MKAMEDVKVSYDRTVR 142
+KA+ED+ V YD T R
Sbjct 842 VKALEDIMVHYDNTTR 857
> ECU03g0290
Length=1599
Score = 198 bits (504), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 96/142 (67%), Positives = 113/142 (79%), Gaps = 0/142 (0%)
Query 1 ARVNKELNAAREKSAKVASEQLDESNNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRI 60
+ +N LN AR+ S A L E+NN+ MVLAGSKGS INISQ+ ACVGQQNVEGKRI
Sbjct 710 SHLNLVLNRARDVSGTSAQRSLSENNNMKTMVLAGSKGSFINISQVTACVGQQNVEGKRI 769
Query 61 PSGFNNRSLPHFSKFDYPPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETG 120
P GF++R+LPHF K DY +SRGFVE+SYLTGL P+E FFHAMGGREG+IDTA KT+ETG
Sbjct 770 PFGFSHRTLPHFVKDDYTGKSRGFVENSYLTGLDPEEFFFHAMGGREGLIDTAIKTAETG 829
Query 121 YIQRRLMKAMEDVKVSYDRTVR 142
YIQRRL+KA+ED V D +VR
Sbjct 830 YIQRRLVKALEDAIVRQDESVR 851
> 7293125
Length=1259
Score = 177 bits (450), Expect = 4e-45, Method: Compositional matrix adjust.
Identities = 79/138 (57%), Positives = 107/138 (77%), Gaps = 0/138 (0%)
Query 5 KELNAAREKSAKVASEQLDESNNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRIPSGF 64
+EL+A RE++AK +L +N+ + M L+GSKGS INISQ++ACVGQQ + GKR+P+GF
Sbjct 628 RELSAIREQAAKTCFAELHPTNSALIMALSGSKGSNINISQMIACVGQQAISGKRVPNGF 687
Query 65 NNRSLPHFSKFDYPPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQR 124
NR+LPHF + P +RGFV++S+ +GL P E FFH M GREG++DTA KT+ETGY+QR
Sbjct 688 ENRALPHFERHSAIPAARGFVQNSFYSGLTPTEFFFHTMAGREGLVDTAVKTAETGYLQR 747
Query 125 RLMKAMEDVKVSYDRTVR 142
RL+K +ED+ V YD TVR
Sbjct 748 RLVKCLEDLVVHYDGTVR 765
> Hs5902062
Length=1391
Score = 173 bits (439), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 78/142 (54%), Positives = 103/142 (72%), Gaps = 0/142 (0%)
Query 1 ARVNKELNAAREKSAKVASEQLDESNNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRI 60
A + KEL+ R+ + +LD+SN+ + M L GSKGS INISQ++ACVGQQ + G R+
Sbjct 750 ALILKELSVIRDHAGSACLRELDKSNSPLTMALCGSKGSFINISQMIACVGQQAISGSRV 809
Query 61 PSGFNNRSLPHFSKFDYPPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETG 120
P GF NRSLPHF K P ++GFV +S+ +GL P E FFH M GREG++DTA KT+ETG
Sbjct 810 PDGFENRSLPHFEKHSKLPAAKGFVANSFYSGLTPTEFFFHTMAGREGLVDTAVKTAETG 869
Query 121 YIQRRLMKAMEDVKVSYDRTVR 142
Y+QRRL+K++ED+ YD TVR
Sbjct 870 YMQRRLVKSLEDLCSQYDLTVR 891
> CE04196
Length=1388
Score = 166 bits (421), Expect = 1e-41, Method: Compositional matrix adjust.
Identities = 75/138 (54%), Positives = 99/138 (71%), Gaps = 0/138 (0%)
Query 5 KELNAAREKSAKVASEQLDESNNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRIPSGF 64
+EL+ R+ + +V L + N + M + GSKGS INISQ++ACVGQQ + G R P GF
Sbjct 750 RELSTIRDHAGQVCLRNLSKYNAPLTMAVCGSKGSFINISQMIACVGQQAISGHRPPDGF 809
Query 65 NNRSLPHFSKFDYPPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQR 124
RSLPHF + PE++GFV +S+ +GL P E FFH MGGREG++DTA KT+ETGY+QR
Sbjct 810 EERSLPHFERKKKTPEAKGFVANSFYSGLTPTEFFFHTMGGREGLVDTAVKTAETGYMQR 869
Query 125 RLMKAMEDVKVSYDRTVR 142
RL+K +ED+ SYD TVR
Sbjct 870 RLVKCLEDLCASYDGTVR 887
> SPBC651.08c
Length=1405
Score = 161 bits (407), Expect = 4e-40, Method: Compositional matrix adjust.
Identities = 71/142 (50%), Positives = 103/142 (72%), Gaps = 0/142 (0%)
Query 1 ARVNKELNAAREKSAKVASEQLDESNNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRI 60
A+++ L+ R+ ++ ++L +N+ + M GSKGS IN+SQ++ACVGQQ + GKR+
Sbjct 744 AKISSTLSKVRDDVGEICMDELGPANSPLIMATCGSKGSKINVSQMVACVGQQIISGKRV 803
Query 61 PSGFNNRSLPHFSKFDYPPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETG 120
P GF +RSLPHF K P ++GFV +S+ +GL P E FHA+ GREG++DTA KT+ETG
Sbjct 804 PDGFQDRSLPHFHKNSKHPLAKGFVSNSFYSGLTPTEFLFHAISGREGLVDTAVKTAETG 863
Query 121 YIQRRLMKAMEDVKVSYDRTVR 142
Y+ RRLMK++ED+ +YD TVR
Sbjct 864 YMSRRLMKSLEDLSSAYDGTVR 885
> At5g60040
Length=1328
Score = 160 bits (404), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 76/142 (53%), Positives = 95/142 (66%), Gaps = 0/142 (0%)
Query 1 ARVNKELNAAREKSAKVASEQLDESNNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRI 60
A + LN RE + K L N+ + M GSKGS INISQ++ACVGQQ V G R
Sbjct 727 AEITGILNTIREATGKACMSGLHWRNSPLIMSQCGSKGSPINISQMVACVGQQTVNGHRA 786
Query 61 PSGFNNRSLPHFSKFDYPPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETG 120
P GF +RSLPHF + P ++GFV +S+ +GL E FFH MGGREG++DTA KT+ TG
Sbjct 787 PDGFIDRSLPHFPRMSKSPAAKGFVANSFYSGLTATEFFFHTMGGREGLVDTAVKTASTG 846
Query 121 YIQRRLMKAMEDVKVSYDRTVR 142
Y+ RRLMKA+ED+ V YD TVR
Sbjct 847 YMSRRLMKALEDLLVHYDNTVR 868
> YOR116c
Length=1460
Score = 160 bits (404), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 72/142 (50%), Positives = 98/142 (69%), Gaps = 0/142 (0%)
Query 1 ARVNKELNAAREKSAKVASEQLDESNNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRI 60
A++ L+ RE+ V +LD N + M GSKGS +N+SQ++A VGQQ + G R+
Sbjct 764 AKIGGLLSKVREEVGDVCINELDNWNAPLIMATCGSKGSTLNVSQMVAVVGQQIISGNRV 823
Query 61 PSGFNNRSLPHFSKFDYPPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETG 120
P GF +RSLPHF K P+S+GFV +S+ +GL P E FHA+ GREG++DTA KT+ETG
Sbjct 824 PDGFQDRSLPHFPKNSKTPQSKGFVRNSFFSGLSPPEFLFHAISGREGLVDTAVKTAETG 883
Query 121 YIQRRLMKAMEDVKVSYDRTVR 142
Y+ RRLMK++ED+ YD TVR
Sbjct 884 YMSRRLMKSLEDLSCQYDNTVR 905
> ECU07g0090
Length=1349
Score = 155 bits (391), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 73/136 (53%), Positives = 96/136 (70%), Gaps = 0/136 (0%)
Query 7 LNAAREKSAKVASEQLDESNNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRIPSGFNN 66
LN RE+ + ++L N+ M GSKGS IN+SQ++ACVGQQ V G RIP+G +
Sbjct 754 LNRIREECGSICIKELGIRNSPNIMQACGSKGSKINVSQMVACVGQQIVSGTRIPNGMDE 813
Query 67 RSLPHFSKFDYPPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRL 126
R LPHF + PES+GFV +S+ GL E FFHA+ GREG++DTA KT+ETGY+QRRL
Sbjct 814 RCLPHFKRGSRTPESKGFVLNSFFDGLTSPEFFFHAVSGREGLVDTAVKTAETGYMQRRL 873
Query 127 MKAMEDVKVSYDRTVR 142
MKA+ED+ + YD +VR
Sbjct 874 MKALEDLSIQYDGSVR 889
> At3g57660
Length=1670
Score = 135 bits (340), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 59/117 (50%), Positives = 85/117 (72%), Gaps = 0/117 (0%)
Query 26 NNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRIPSGFNNRSLPHFSKFDYPPESRGFV 85
N I M ++G+KGS +N QI + +GQQ++EGKR+P + ++LP F +D+ P + GF+
Sbjct 936 NCISLMTISGAKGSKVNFQQISSHLGQQDLEGKRVPRMVSGKTLPCFHPWDWSPRAGGFI 995
Query 86 EHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRLMKAMEDVKVSYDRTVR 142
+L+GL PQE +FH M GREG++DTA KTS +GY+QR LMK +E +KV+YD TVR
Sbjct 996 SDRFLSGLRPQEYYFHCMAGREGLVDTAVKTSRSGYLQRCLMKNLESLKVNYDCTVR 1052
> 7303170
Length=1642
Score = 135 bits (339), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 61/118 (51%), Positives = 85/118 (72%), Gaps = 0/118 (0%)
Query 25 SNNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRIPSGFNNRSLPHFSKFDYPPESRGF 84
SNN+ MVL+G+KGS++N QI +GQ +EGKR P + +SLP F+ F+ P+S GF
Sbjct 869 SNNLQLMVLSGAKGSMVNTMQISCLLGQIELEGKRPPLMISGKSLPSFTSFETSPKSGGF 928
Query 85 VEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRLMKAMEDVKVSYDRTVR 142
++ ++TG+ PQ+ FFH M GREG+IDTA KTS +GY+QR L+K +E + V YD TVR
Sbjct 929 IDGRFMTGIQPQDFFFHCMAGREGLIDTAVKTSRSGYLQRCLIKHLEGLSVHYDLTVR 986
> ECU04g1400
Length=1395
Score = 130 bits (327), Expect = 9e-31, Method: Compositional matrix adjust.
Identities = 62/119 (52%), Positives = 86/119 (72%), Gaps = 2/119 (1%)
Query 26 NNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRIPSGFNNRSLPHFSKFDYPPESRGFV 85
NN+ ++ GSKGSI+N+SQI +GQQ +EG+R+P + ++LP F+ D P S G++
Sbjct 761 NNMELIIATGSKGSIVNLSQISGALGQQELEGQRVPIMASGKTLPCFAALDPSPSSGGYI 820
Query 86 EHSYLTGL-LPQELFFHAMGGREGIIDTACKTSETGYIQRRLMKAMEDVKVSYDRTVRI 143
H +LTG+ LPQ FFH M GREG+IDTA KT+ +GY+QR L+K ME+ KV YD +VRI
Sbjct 821 YHRFLTGIDLPQ-YFFHCMAGREGLIDTAIKTANSGYLQRCLIKHMEEAKVEYDMSVRI 878
> CE22157
Length=1737
Score = 130 bits (326), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 58/117 (49%), Positives = 81/117 (69%), Gaps = 0/117 (0%)
Query 26 NNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRIPSGFNNRSLPHFSKFDYPPESRGFV 85
N + M+ +G+KGS +N QI C+GQ +EGKR+ R+LP F FD P + G++
Sbjct 928 NALQLMIQSGAKGSAVNAIQISGCLGQIELEGKRMAVTIAGRTLPSFRCFDPSPRAGGYI 987
Query 86 EHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRLMKAMEDVKVSYDRTVR 142
+ +LTG+ PQELFFH M GREG+IDTA KTS +GY+QR ++K +E ++V YD TVR
Sbjct 988 DQRFLTGMNPQELFFHTMAGREGLIDTAVKTSRSGYLQRCIIKHLEGIRVHYDSTVR 1044
> SPBC4C3.05c
Length=1689
Score = 129 bits (325), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 58/117 (49%), Positives = 83/117 (70%), Gaps = 0/117 (0%)
Query 26 NNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRIPSGFNNRSLPHFSKFDYPPESRGFV 85
N++ M ++G+KGS +N+SQI +GQQ +EG+R+P + +SLP F ++ +S GF+
Sbjct 936 NHMQTMTVSGAKGSNVNVSQISCLLGQQELEGRRVPLMVSGKSLPSFVPYETSAKSGGFI 995
Query 86 EHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRLMKAMEDVKVSYDRTVR 142
+LTG+ PQE +FH M GREG+IDTA KTS +GY+QR LMK +E + V YD TVR
Sbjct 996 ASRFLTGIAPQEYYFHCMAGREGLIDTAVKTSRSGYLQRCLMKHLEGLCVQYDHTVR 1052
> YOR341w
Length=1664
Score = 127 bits (320), Expect = 6e-30, Method: Compositional matrix adjust.
Identities = 56/117 (47%), Positives = 84/117 (71%), Gaps = 0/117 (0%)
Query 26 NNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRIPSGFNNRSLPHFSKFDYPPESRGFV 85
N++ AM L+G+KGS +N+SQIM +GQQ +EG+R+P + ++LP F ++ + G+V
Sbjct 923 NSMQAMALSGAKGSNVNVSQIMCLLGQQALEGRRVPVMVSGKTLPSFKPYETDAMAGGYV 982
Query 86 EHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRLMKAMEDVKVSYDRTVR 142
+ + +G+ PQE +FH M GREG+IDTA KTS +GY+QR L K +E V VSYD ++R
Sbjct 983 KGRFYSGIKPQEYYFHCMAGREGLIDTAVKTSRSGYLQRCLTKQLEGVHVSYDNSIR 1039
> Hs7661686
Length=1717
Score = 118 bits (296), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 54/117 (46%), Positives = 77/117 (65%), Gaps = 0/117 (0%)
Query 26 NNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRIPSGFNNRSLPHFSKFDYPPESRGFV 85
N + MV +G+KGS +N QI +GQ +EG+ P + +SLP F +++ P + GFV
Sbjct 892 NTLQLMVQSGAKGSTVNTMQISCLLGQIELEGRSTPLMASGKSLPCFEPYEFTPRAGGFV 951
Query 86 EHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRLMKAMEDVKVSYDRTVR 142
+LTG+ P E FFH M GREG++DTA KTS +GY+QR ++K +E + V YD TVR
Sbjct 952 TGRFLTGIKPPEFFFHCMAGREGLVDTAVKTSRSGYLQRCIIKHLEGLVVQYDLTVR 1008
> At1g63020_1
Length=1316
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 43/136 (31%), Positives = 65/136 (47%), Gaps = 20/136 (14%)
Query 23 DESNNIMAMVLAGSKGSIINISQIMACVGQQN--------VEGKRIPSGFNNRSLP---- 70
D+SN+ + M AGSKG+I + Q C+G QN + + +N+ + P
Sbjct 720 DQSNSFLIMSKAGSKGNIGKLVQHSMCIGLQNSAVSLSFGFPRELTCAAWNDPNSPLRGA 779
Query 71 ----HFSKFDYPPESRGFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRL 126
+ Y P G +E+S+LTGL P E F H++ R+ + G + RRL
Sbjct 780 KGKDSTTTESYVP--YGVIENSFLTGLNPLESFVHSVTSRDSSF--SGNADLPGTLSRRL 835
Query 127 MKAMEDVKVSYDRTVR 142
M M D+ +YD TVR
Sbjct 836 MFFMRDIYAAYDGTVR 851
> AtCh013
Length=1376
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 31/113 (27%), Positives = 52/113 (46%), Gaps = 20/113 (17%)
Query 23 DESNNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRIPSGFNNRSLPHFSKFDYPPESR 82
D N + M +G++G N SQ+ VG + + S P D P
Sbjct 123 DPFNPVHMMSFSGARG---NASQVHQLVGMRGL-----------MSDPQGQMIDLP---- 164
Query 83 GFVEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRRLMKAMEDVKV 135
++ + GL E G R+G++DTA +TS+ GY+ RRL++ ++ + V
Sbjct 165 --IQSNLREGLSLTEYIISCYGARKGVVDTAVRTSDAGYLTRRLVEVVQHIVV 215
> CE28721
Length=403
Score = 29.6 bits (65), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 18/69 (26%), Positives = 32/69 (46%), Gaps = 10/69 (14%)
Query 10 AREKSAKVASEQLDESNNIMAMVLAGSKGSIINISQIMACVGQQNVEGKRIPSGFNNR-- 67
R++ A + +D N+M ++ +K ++ N+ + C GQQ E IP + R
Sbjct 3 TRDEDENWAKKAID---NLMKKLIKHNKQALENLEFALRCQGQQKTECVTIPRSLDGRLQ 59
Query 68 -----SLPH 71
+LPH
Sbjct 60 ISHRKALPH 68
> Hs11321632
Length=709
Score = 29.6 bits (65), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 17/28 (60%), Gaps = 1/28 (3%)
Query 73 SKFDYPPESRGFV-EHSYLTGLLPQELF 99
S+ DYPP + F EH +T L PQ+L
Sbjct 87 SEIDYPPFEKNFYNEHEEITNLTPQQLI 114
> At3g16370
Length=353
Score = 29.3 bits (64), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 11/23 (47%), Positives = 12/23 (52%), Gaps = 0/23 (0%)
Query 65 NNRSLPHFSKFDYPPESRGFVEH 87
NN LP + DYPP R F H
Sbjct 43 NNNYLPTLFRADYPPYGRDFANH 65
> SPAC1952.09c
Length=521
Score = 28.5 bits (62), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 18/41 (43%), Positives = 22/41 (53%), Gaps = 4/41 (9%)
Query 85 VEHSYLTGLLPQELFFHAMGGREGIIDTACKTSETGYIQRR 125
VE + L G L +LF A GG DT CK +E I+RR
Sbjct 57 VEENNLQGKLRFKLFVGASGGA----DTECKWAELNMIERR 93
> Hs20545656
Length=284
Score = 27.7 bits (60), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 22/93 (23%), Positives = 46/93 (49%), Gaps = 7/93 (7%)
Query 1 ARVNKELNAAREKSAKVASEQLDESNNIMA---MVLAGSKGSI-INISQIMACVGQQN-- 54
AR + ++ A + + L + ++A VL G KG+ + + +++ VGQ+
Sbjct 50 ARQRGTTSRRKDAVACILRQALPCGSEVLAAIHCVLPGCKGADGLGVGEVVKFVGQRRYG 109
Query 55 -VEGKRIPSGFNNRSLPHFSKFDYPPESRGFVE 86
+ GKR+ S F + LP ++ + +GF++
Sbjct 110 VMTGKRMLSIFTVKKLPISERYTLGLQQQGFIK 142
Lambda K H
0.318 0.132 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1712413322
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40