bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1555_orf1
Length=168
Score E
Sequences producing significant alignments: (Bits) Value
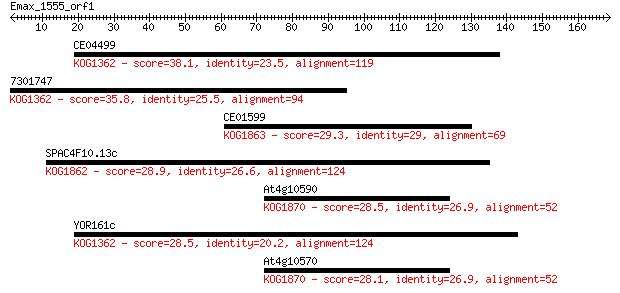
CE04499 38.1 0.009
7301747 35.8 0.037
CE01599 29.3 3.9
SPAC4F10.13c 28.9 5.6
At4g10590 28.5 6.6
YOR161c 28.5 7.1
At4g10570 28.1 8.0
> CE04499
Length=771
Score = 38.1 bits (87), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 28/119 (23%), Positives = 50/119 (42%), Gaps = 2/119 (1%)
Query 19 HHLGSFAVGGLVMGVTRPIRLLFAWAATKHLASTEDSPVVQSLRDSFRNIISPLAFVVDR 78
++LGS A G L++ + + IR+L + H + V+ + L
Sbjct 564 YNLGSIAFGSLIIAIVKIIRVLLEY--IDHKLGKSQNKAVKWFLMCLKCCFWCLEVFFKF 621
Query 79 FTSSGYIEMSISSKPFFPAMDKSHTRLNQARSPATYLHGVVAIASAIGSLFISLLIGIM 137
T + YI ++I K FF + S + + +H V I +G I+L +GI+
Sbjct 622 LTKNAYIMIAIYGKNFFSSAKDSFLLITRNIVRTVVVHKVAGILLFLGKSMITLGMGIL 680
> 7301747
Length=796
Score = 35.8 bits (81), Expect = 0.037, Method: Composition-based stats.
Identities = 24/94 (25%), Positives = 43/94 (45%), Gaps = 2/94 (2%)
Query 1 ERDAGWFSPLVAIGLGSIHHLGSFAVGGLVMGVTRPIRLLFAWAATKHLASTEDSPVVQS 60
+RD +F+ A +++HLG+ A G L++ + R IRL+ + K D+ V ++
Sbjct 573 KRDVPYFTLTRAFFQTAVYHLGTVAFGSLILAIVRLIRLVLEYIHEK--LKKYDNAVTRA 630
Query 61 LRDSFRNIISPLAFVVDRFTSSGYIEMSISSKPF 94
+ R L + + YI +I K F
Sbjct 631 ILCCMRCFFWLLETFLKFLNRNAYIMCAIHGKNF 664
> CE01599
Length=1302
Score = 29.3 bits (64), Expect = 3.9, Method: Composition-based stats.
Identities = 20/72 (27%), Positives = 39/72 (54%), Gaps = 5/72 (6%)
Query 61 LRDSFRNIISPLAF---VVDRFTSSGYIEMSISSKPFFPAMDKSHTRLNQARSPATYLHG 117
L++SF NI S + + V + T S ++ ++ + P F + ++ L+ A AT+ HG
Sbjct 444 LKESFNNIFSSVCYTESVAEDGTVS--VKSNVRNCPQFFQLQVTYGNLHDALEAATFDHG 501
Query 118 VVAIASAIGSLF 129
+ AS + +L+
Sbjct 502 LGNTASHVRNLY 513
> SPAC4F10.13c
Length=992
Score = 28.9 bits (63), Expect = 5.6, Method: Composition-based stats.
Identities = 33/128 (25%), Positives = 56/128 (43%), Gaps = 20/128 (15%)
Query 11 VAIGLGSIHHLGSFAVGGLVMGVTRPIRLLFAWAATKHLASTEDSPVVQSLRDSFRNIIS 70
VA +G + +L F+ L G +R + + PV S+ S R+ S
Sbjct 276 VASPVGQVDNLADFSQSPLRRGPSR-------------FPTNSNVPVGNSM--SIRDTDS 320
Query 71 PLAFVVDRFTSSGYIEMSISSKPFFP-AMDKSHTRLNQ---ARSPATYLHGVVAIASAIG 126
PL +VD+ + I+ + +S+P P A + H+ +N A SP+T + A +
Sbjct 321 PLNILVDKAKAKASIKEN-ASQPVAPSASQREHSAVNSPAAAMSPSTAMFSSEAFPQHLA 379
Query 127 SLFISLLI 134
SL L+
Sbjct 380 SLIPPALL 387
> At4g10590
Length=937
Score = 28.5 bits (62), Expect = 6.6, Method: Composition-based stats.
Identities = 14/52 (26%), Positives = 29/52 (55%), Gaps = 0/52 (0%)
Query 72 LAFVVDRFTSSGYIEMSISSKPFFPAMDKSHTRLNQARSPATYLHGVVAIAS 123
L F + RFT S Y++ I + FP D ++ + ++ +YL+ + A+++
Sbjct 819 LVFHLKRFTYSRYLKNKIDTFVNFPVHDLDLSKYVKNKNDQSYLYELYAVSN 870
> YOR161c
Length=539
Score = 28.5 bits (62), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 25/124 (20%), Positives = 50/124 (40%), Gaps = 0/124 (0%)
Query 19 HHLGSFAVGGLVMGVTRPIRLLFAWAATKHLASTEDSPVVQSLRDSFRNIISPLAFVVDR 78
+ GS G L++ + +R + +S +Q L F II L ++ +
Sbjct 335 YSFGSICFGSLLVALIDLLRQILQMIRHDVTSSGGGQIAIQILFMVFDWIIGFLKWLAEY 394
Query 79 FTSSGYIEMSISSKPFFPAMDKSHTRLNQARSPATYLHGVVAIASAIGSLFISLLIGIMI 138
F Y +++ KP+ A ++ L + A ++ IA + S+F S + +
Sbjct 395 FNHYAYSFIALYGKPYLRAAKETWYMLREKGMDALINDNLINIALGLFSMFASYMTALFT 454
Query 139 YSTL 142
+ L
Sbjct 455 FLYL 458
> At4g10570
Length=928
Score = 28.1 bits (61), Expect = 8.0, Method: Composition-based stats.
Identities = 14/52 (26%), Positives = 29/52 (55%), Gaps = 0/52 (0%)
Query 72 LAFVVDRFTSSGYIEMSISSKPFFPAMDKSHTRLNQARSPATYLHGVVAIAS 123
L F + RFT S Y++ I + FP D ++ + ++ +YL+ + A+++
Sbjct 798 LVFHLKRFTYSRYLKNKIDTFVNFPVHDLDLSKYVKNKNGQSYLYELYAVSN 849
Lambda K H
0.323 0.134 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2418402922
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40