bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1536_orf1
Length=164
Score E
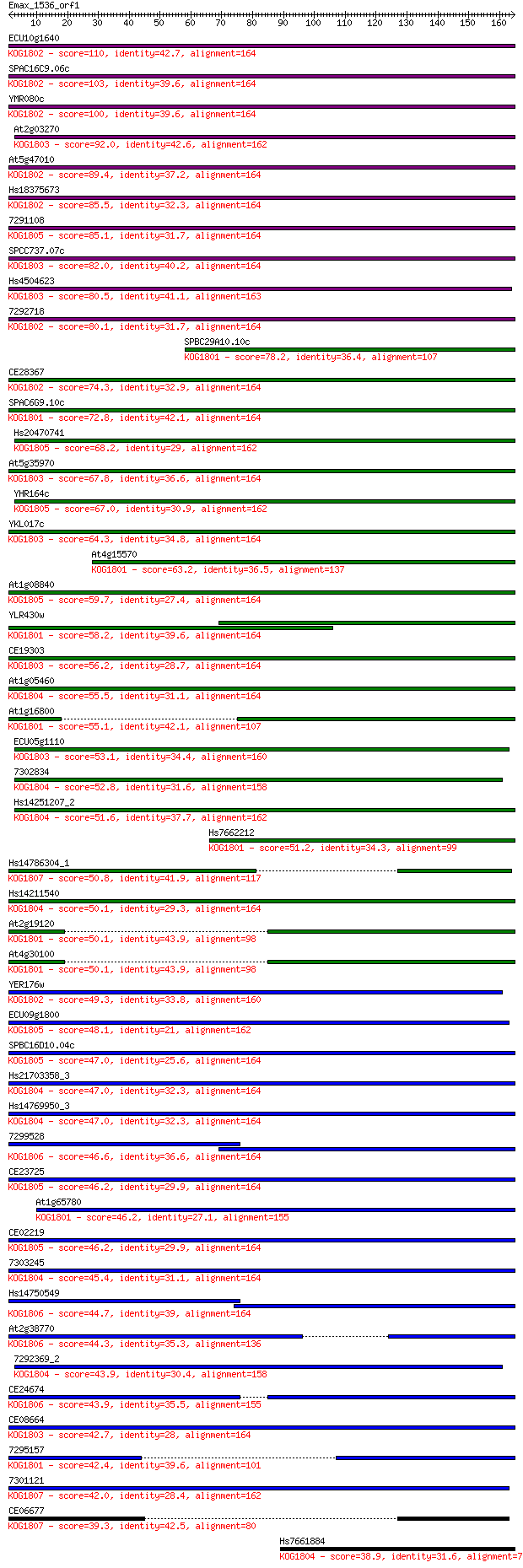
Sequences producing significant alignments: (Bits) Value
ECU10g1640 110 1e-24
SPAC16C9.06c 103 2e-22
YMR080c 100 1e-21
At2g03270 92.0 5e-19
At5g47010 89.4 3e-18
Hs18375673 85.5 4e-17
7291108 85.1 6e-17
SPCC737.07c 82.0 4e-16
Hs4504623 80.5 1e-15
7292718 80.1 2e-15
SPBC29A10.10c 78.2 6e-15
CE28367 74.3 9e-14
SPAC6G9.10c 72.8 3e-13
Hs20470741 68.2 8e-12
At5g35970 67.8 1e-11
YHR164c 67.0 2e-11
YKL017c 64.3 9e-11
At4g15570 63.2 2e-10
At1g08840 59.7 3e-09
YLR430w 58.2 8e-09
CE19303 56.2 2e-08
At1g05460 55.5 5e-08
At1g16800 55.1 6e-08
ECU05g1110 53.1 2e-07
7302834 52.8 3e-07
Hs14251207_2 51.6 6e-07
Hs7662212 51.2 9e-07
Hs14786304_1 50.8 1e-06
Hs14211540 50.1 2e-06
At2g19120 50.1 2e-06
At4g30100 50.1 2e-06
YER176w 49.3 3e-06
ECU09g1800 48.1 7e-06
SPBC16D10.04c 47.0 2e-05
Hs21703358_3 47.0 2e-05
Hs14769950_3 47.0 2e-05
7299528 46.6 2e-05
CE23725 46.2 3e-05
At1g65780 46.2 3e-05
CE02219 46.2 3e-05
7303245 45.4 5e-05
Hs14750549 44.7 9e-05
At2g38770 44.3 1e-04
7292369_2 43.9 1e-04
CE24674 43.9 2e-04
CE08664 42.7 3e-04
7295157 42.4 4e-04
7301121 42.0 6e-04
CE06677 39.3 0.004
Hs7661884 38.9 0.005
> ECU10g1640
Length=782
Score = 110 bits (275), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 70/174 (40%), Positives = 102/174 (58%), Gaps = 11/174 (6%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGS-- 58
++GPPGTGKT V+ AI+ R + K+L VA SN A D L + G+R +RV S
Sbjct 364 IQGPPGTGKTLVSSAIVYNLVR-HYGGKVLVVAPSNTAVDQLTLKIHKTGLRVLRVMSRR 422
Query 59 --GSESDLKEESI-QDLARYRDLLRIRDKNSGEARSLRMLLVREAVRKY-----NVIIAT 110
+SD+ S+ ++L ++ + +D++ S+ V E+++K VI T
Sbjct 423 REYGQSDVSFLSLHENLRELQEGRKRKDEDHSRYDSIYNDEVNESLKKQLLNQAEVITCT 482
Query 111 CVGSGHEMFDDVTFERVIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLPPTI 164
CV SG +MF+ F V+IDE QS EP +LIPL +GC+ +VL+GDHKQL PTI
Sbjct 483 CVTSGQKMFNRFKFHCVLIDEAVQSTEPLSLIPLVYGCKKLVLVGDHKQLGPTI 536
> SPAC16C9.06c
Length=925
Score = 103 bits (257), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 65/188 (34%), Positives = 100/188 (53%), Gaps = 25/188 (13%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKK-----ILAVADSNVAADNLMEGLSNRGIRSVR 55
++GPPGTGKT + +++ + K+ +L A SNVA D L E + G+R VR
Sbjct 412 IQGPPGTGKTVTSASVVYHLATMQSRKRKSHSPVLVCAPSNVAVDQLAEKIHRTGLRVVR 471
Query 56 VGSGSESDLKEESIQDLARYRD------------LLRIRDKNS----GEARSLRMLLV-- 97
V + S D+ E S+ L+ + LL++R +N+ + + LR+L+
Sbjct 472 VAAKSREDI-ESSVSFLSLHEQIKNYKFNPELQRLLKLRSENNELSIQDEKKLRILVAAA 530
Query 98 -REAVRKYNVIIATCVGSGHEMFDDVTFERVIIDECAQSIEPSNLIPLGHGCRSVVLIGD 156
+E +R +VI TCVG+G F V+IDE Q+ EP +IPL G + VVL+GD
Sbjct 531 EKELLRAAHVICCTCVGAGDRRISKYKFRSVLIDEATQASEPECMIPLVLGAKQVVLVGD 590
Query 157 HKQLPPTI 164
H+QL P +
Sbjct 591 HQQLGPVV 598
> YMR080c
Length=971
Score = 100 bits (250), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 65/182 (35%), Positives = 104/182 (57%), Gaps = 21/182 (11%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGSGS 60
++GPPGTGKT + I+ +I+ +IL A SNVA D+L L + G++ VR+ + S
Sbjct 428 IQGPPGTGKTVTSATIVYHLSKIH-KDRILVCAPSNVAVDHLAAKLRDLGLKVVRLTAKS 486
Query 61 ESDLKEESIQDLARY-----------RDLLRIRDK-----NSGEARSLRMLLVREA--VR 102
D+ E S+ +LA + ++LL+++D+ S R ++++ EA +
Sbjct 487 REDV-ESSVSNLALHNLVGRGAKGELKNLLKLKDEVGELSASDTKRFVKLVRKTEAEILN 545
Query 103 KYNVIIATCVGSGHEMFDDVTFERVIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLPP 162
K +V+ TCVG+G + D F V+IDE Q+ EP LIP+ G + V+L+GDH+QL P
Sbjct 546 KADVVCCTCVGAGDKRLD-TKFRTVLIDESTQASEPECLIPIVKGAKQVILVGDHQQLGP 604
Query 163 TI 164
I
Sbjct 605 VI 606
> At2g03270
Length=635
Score = 92.0 bits (227), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 69/197 (35%), Positives = 96/197 (48%), Gaps = 41/197 (20%)
Query 3 GPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVG----- 57
GPPGTGKT I+ Q + KILA A SN+A DN++E L ++ VRVG
Sbjct 208 GPPGTGKTTTVVEIV--LQEVKRGSKILACAASNIAVDNIVERLVPHKVKLVRVGHPARL 265
Query 58 ------------------SGSESDLKEESIQDLARYRDLLRIRDKNS------------G 87
SG +D+++E A LL+ +DKN+
Sbjct 266 LPQVLDSALDAQVLKGDNSGLANDIRKEMK---ALNGKLLKAKDKNTRRLIQKELRTLGK 322
Query 88 EARSLRMLLVREAVRKYNVIIATCVGSGHEMFDDVTFERVIIDECAQSIEPSNLIPLGHG 147
E R + L V + ++ +VI+ T G+ D+ TF+ VIIDE AQ++E + I L G
Sbjct 323 EERKRQQLAVSDVIKNADVILTTLTGALTRKLDNRTFDLVIIDEGAQALEVACWIALLKG 382
Query 148 CRSVVLIGDHKQLPPTI 164
R +L GDH QLPPTI
Sbjct 383 SR-CILAGDHLQLPPTI 398
> At5g47010
Length=1235
Score = 89.4 bits (220), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 61/184 (33%), Positives = 96/184 (52%), Gaps = 21/184 (11%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGSGS 60
++GPPGTGKT + AI+ + ++L A SNVA D L E +S G++ VR+ + S
Sbjct 505 IQGPPGTGKTVTSAAIVYHMAKQG-QGQVLVCAPSNVAVDQLAEKISATGLKVVRLCAKS 563
Query 61 ----ESDLKEESIQDLARYRD---------LLRIRDK-------NSGEARSLRMLLVREA 100
S ++ ++ R+ D L +++D+ + + ++L+ RE
Sbjct 564 REAVSSPVEYLTLHYQVRHLDTSEKSELHKLQQLKDEQGELSSSDEKKYKNLKRATEREI 623
Query 101 VRKYNVIIATCVGSGHEMFDDVTFERVIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQL 160
+ +VI TCVG+ + F +V+IDE Q+ EP LIPL G + VVL+GDH QL
Sbjct 624 TQSADVICCTCVGAADLRLSNFRFRQVLIDESTQATEPECLIPLVLGVKQVVLVGDHCQL 683
Query 161 PPTI 164
P I
Sbjct 684 GPVI 687
> Hs18375673
Length=1118
Score = 85.5 bits (210), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 53/183 (28%), Positives = 91/183 (49%), Gaps = 21/183 (11%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGSGS 60
++GPPGTGKT + I+ R +L A SN+A D L E + G++ VR+ + S
Sbjct 490 IQGPPGTGKTVTSATIVYHLARQG-NGPVLVCAPSNIAVDQLTEKIHQTGLKVVRLCAKS 548
Query 61 ESDLKE-----------ESIQDLARYRDLLRIRDKNSGE--------ARSLRMLLVREAV 101
+ ++ + + L +++D+ +GE R+L+ RE +
Sbjct 549 REAIDSPVSFLALHNQIRNMDSMPELQKLQQLKDE-TGELSSADEKRYRALKRTAERELL 607
Query 102 RKYNVIIATCVGSGHEMFDDVTFERVIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLP 161
+VI TCVG+G + F ++IDE Q+ EP ++P+ G + ++L+GDH QL
Sbjct 608 MNADVICCTCVGAGDPRLAKMQFRSILIDESTQATEPECMVPVVLGAKQLILVGDHCQLG 667
Query 162 PTI 164
P +
Sbjct 668 PVV 670
> 7291108
Length=1119
Score = 85.1 bits (209), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 52/164 (31%), Positives = 85/164 (51%), Gaps = 16/164 (9%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGSGS 60
+KG PGTGKT A++ + K +L A ++ A DNL+ L G+ +R+GSGS
Sbjct 676 IKGLPGTGKTQTLVALVRLLHLLG--KSVLITAQTHSAVDNLIMRLLPFGLPMMRLGSGS 733
Query 61 ESDLKEESIQDLARYRDLLRIRDKNSGEARSLRMLLVREAVRKYNVIIATCVGSGHEMFD 120
+ E I + +D + + + +A+ + +++ TC+G GH +F
Sbjct 734 RIHKQLEEISEERLTKDCKTVEE-------------LEKALGQPSIVGVTCLGCGHPLFQ 780
Query 121 DVTFERVIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLPPTI 164
F+ I+DE Q ++P+ L PL H C VL+GD +QLPP +
Sbjct 781 RRQFDYCIVDEATQVLQPTVLRPLIH-CSKFVLVGDPEQLPPIV 823
> SPCC737.07c
Length=660
Score = 82.0 bits (201), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 66/196 (33%), Positives = 94/196 (47%), Gaps = 35/196 (17%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGSGS 60
+ GPPGTGKTH II Q + K+IL SN+A DN+++ LS+ GI VR+G +
Sbjct 230 IHGPPGTGKTHTLVEIIQ--QLVLRNKRILVCGASNLAVDNIVDRLSSSGIPMVRLGHPA 287
Query 61 E--SDLKEESIQDLARYRD---------------LLRI-RDKNSGEARSLRMLL--VREA 100
+ + S+ L+R D L +I + KN E R + + +R+
Sbjct 288 RLLPSILDHSLDVLSRTGDNGDVIRGISEDIDVCLSKITKTKNGRERREIYKNIRELRKD 347
Query 101 VRKY------------NVIIATCVGSGHEMFDDVTFERVIIDECAQSIEPSNLIPLGHGC 148
RKY V+ T G+G F+ VIIDE +Q++EP IPL G
Sbjct 348 YRKYEAKTVANIVSASKVVFCTLHGAGSRQLKGQRFDAVIIDEASQALEPQCWIPL-LGM 406
Query 149 RSVVLIGDHKQLPPTI 164
V+L GDH QL P +
Sbjct 407 NKVILAGDHMQLSPNV 422
> Hs4504623
Length=993
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 67/199 (33%), Positives = 100/199 (50%), Gaps = 39/199 (19%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVG--- 57
+ GPPGTGKT II Q + K+L A SN+A DNL+E L+ R +R+G
Sbjct 212 IHGPPGTGKTTTVVEII--LQAVKQGLKVLCCAPSNIAVDNLVERLALCKQRILRLGHPA 269
Query 58 ----SGSESDL-----KEESIQDLARYR---DLLRIRDKNSGEARS----------LRML 95
S + L + +S Q++A R D + +++K + + R LR
Sbjct 270 RLLESVQQHSLDAVLARSDSAQNVADIRKDIDQVFVKNKKTQDKREKSNFRNEIKLLRKE 329
Query 96 L-------VREAVRKYNVIIATCVGSGHE----MFDDVTFERVIIDECAQSIEPSNLIPL 144
L + E++ NV++AT G+ + + + F+ V+IDECAQ++E S IPL
Sbjct 330 LKEREEAAMLESLTSANVVLATNTGASADGPLKLLPESYFDVVVIDECAQALEASCWIPL 389
Query 145 GHGCRSVVLIGDHKQLPPT 163
R +L GDHKQLPPT
Sbjct 390 LKA-RKCILAGDHKQLPPT 407
> 7292718
Length=685
Score = 80.1 bits (196), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 52/183 (28%), Positives = 91/183 (49%), Gaps = 21/183 (11%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGSGS 60
++GPPGTGKT + I+ +++ +L A SN A D L E + ++ VRV + S
Sbjct 471 IQGPPGTGKTVTSATIVYQLVKLH-GGTVLVCAPSNTAVDQLTEKIHRTNLKVVRVCAKS 529
Query 61 ESDLKE-----------ESIQDLARYRDLLRIRDKNSGE--------ARSLRMLLVREAV 101
+ +++ + + L +++D+ +GE R+L+ + +
Sbjct 530 REAIDSPVSFLALHNQIRNMETNSELKKLQQLKDE-TGELSSADEKRYRNLKRAAENQLL 588
Query 102 RKYNVIIATCVGSGHEMFDDVTFERVIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLP 161
+VI TCVG+G V F ++IDE QS EP ++P+ G + ++L+GDH QL
Sbjct 589 EAADVICCTCVGAGDGRLSRVKFTSILIDESMQSTEPECMVPVVLGAKQLILVGDHCQLG 648
Query 162 PTI 164
P +
Sbjct 649 PVV 651
> SPBC29A10.10c
Length=1944
Score = 78.2 bits (191), Expect = 6e-15, Method: Composition-based stats.
Identities = 39/117 (33%), Positives = 73/117 (62%), Gaps = 10/117 (8%)
Query 58 SGSESDLKEESIQDLARYRDLLR-----IRDKNSGEARSLRML---LVREAVRKYNVIIA 109
+G S + E ++++ + +++L +R++ R+L +L + + +++ +++ A
Sbjct 1434 TGKNSSILEAQLREITKQKNMLEQSLDDMRERQRSTNRNLDVLKKQIQNQLLQEADIVCA 1493
Query 110 TCVGSGHEMFDD--VTFERVIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLPPTI 164
T SGHE+ + +TF VIIDE AQ++E S++IPL +GC S V++GD QLPPT+
Sbjct 1494 TLSASGHELLLNAGLTFRTVIIDEAAQAVELSSIIPLKYGCESCVMVGDPNQLPPTV 1550
> CE28367
Length=1069
Score = 74.3 bits (181), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 54/183 (29%), Positives = 95/183 (51%), Gaps = 22/183 (12%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTK-KILAVADSNVAADNLMEGLSNRGIRSVRVGS- 58
++GPPGTGKT V+ I+ + + T+ +L + SN+A D+L E + G++ VR+ +
Sbjct 465 IQGPPGTGKTVVSATIV--YHLVQKTEGNVLVCSPSNIAVDHLAEKIHKTGLKVVRLCAR 522
Query 59 ---GSESDLKEESIQDL------ARYRDLLRIRDKNSGEAR---SLRMLLVR-----EAV 101
SE+ + ++Q A + L++++D+ +GE LR + ++ E +
Sbjct 523 SREHSETTVPYLTLQHQLKVMGGAELQKLIQLKDE-AGELEFKDDLRYMQLKRVKEHELL 581
Query 102 RKYNVIIATCVGSGHEMFDDVTFERVIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLP 161
+VI TC + + V+IDE Q+ EP L+ + G R +VL+GDH QL
Sbjct 582 AAADVICCTCSSAADARLSKIRTRTVLIDESTQATEPEILVSIMRGVRQLVLVGDHCQLG 641
Query 162 PTI 164
P +
Sbjct 642 PVV 644
> SPAC6G9.10c
Length=1687
Score = 72.8 bits (177), Expect = 3e-13, Method: Composition-based stats.
Identities = 69/264 (26%), Positives = 101/264 (38%), Gaps = 101/264 (38%)
Query 1 VKGPPGTGKTHVACAIIDAW----------------QRINPTKKILAVADSNVAADNLME 44
++GPPGTGKT II A + ++IL A SN A D ++
Sbjct 1150 IQGPPGTGKTKTIIGIISALLVDLSRYHITRPNQQSKSTESKQQILLCAPSNAAVDEVLL 1209
Query 45 GLSNRGI----------RSVRVGSG-----SESDLKEE------------------SIQD 71
L RG R VR+G+ S DL E S+Q+
Sbjct 1210 RL-KRGFLLENGEKYIPRVVRIGNPETINVSVRDLSLEYQTEKQLLEVNQGAIDLGSLQE 1268
Query 72 LARYRDLL---------------------------------RIRDKNSGEARS------- 91
L R+RD +I +KN E +
Sbjct 1269 LTRWRDTFYDCIQKIEELEKQIDVARDVAEDTKSLGKELQNKINEKNLAEQKVEELQSQS 1328
Query 92 ---------LRMLLVREAVRKYNVIIATCVGSGHEMF--DDVTFERVIIDECAQSIEPSN 140
LR + +++ +V+ AT GSGH++ + F VIIDE AQ++E
Sbjct 1329 FTKNKEVDLLRKKAQKAILKQADVVCATLSGSGHDLVAHSSLNFSTVIIDEAAQAVELDT 1388
Query 141 LIPLGHGCRSVVLIGDHKQLPPTI 164
+IPL +G + +L+GD QLPPT+
Sbjct 1389 IIPLRYGAKKCILVGDPNQLPPTV 1412
> Hs20470741
Length=1060
Score = 68.2 bits (165), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 47/165 (28%), Positives = 83/165 (50%), Gaps = 20/165 (12%)
Query 3 GPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGSGSES 62
G PGTGKT C ++ + +L + ++ A DN++ L+ I +R+G +
Sbjct 648 GMPGTGKTTTICTLVRILYACGFS--VLLTSYTHSAVDNILLKLAKFKIGFLRLGQIQKV 705
Query 63 D--LKEESIQDLARYRDLLRIRDKNSGEARSLRMLLVREAVRKYNVIIAT-CVGSGHEMF 119
+++ + Q++ R ++S++ L + E + +I+AT C+G H +F
Sbjct 706 HPAIQQFTEQEICR--------------SKSIKSLALLEELYNSQLIVATTCMGINHPIF 751
Query 120 DDVTFERVIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLPPTI 164
F+ I+DE +Q +P L PL R VL+GDH+QLPP +
Sbjct 752 SRKIFDFCIVDEASQISQPICLGPLFFS-RRFVLVGDHQQLPPLV 795
> At5g35970
Length=750
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 60/199 (30%), Positives = 92/199 (46%), Gaps = 40/199 (20%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGSGS 60
V+GPPGTGKT + +I + +++L A +N A DN++E L + G+ VRVG+ +
Sbjct 297 VQGPPGTGKTGMLKEVITL--AVQQGERVLVTAPTNAAVDNMVEKLLHLGLNIVRVGNPA 354
Query 61 -------ESDLKEESIQDLARYRDLLRIRDKNSGEARSLRMLL--------VREAVRKY- 104
L E LA +R + K S + LR L +R+ +++
Sbjct 355 RISSAVASKSLGEIVNSKLASFR--AELERKKSDLRKDLRQCLRDDVLAAGIRQLLKQLG 412
Query 105 ------------------NVIIATCVGSGHEMFDDV-TFERVIIDECAQSIEPSNLIPLG 145
V+ AT +G+ + + TF+ V+IDE QSIEPS IP+
Sbjct 413 KTLKKKEKETVKEILSNAQVVFATNIGAADPLIRRLETFDLVVIDEAGQSIEPSCWIPIL 472
Query 146 HGCRSVVLIGDHKQLPPTI 164
G R +L GD QL P +
Sbjct 473 QGKR-CILSGDPCQLAPVV 490
> YHR164c
Length=1522
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 50/164 (30%), Positives = 81/164 (49%), Gaps = 20/164 (12%)
Query 3 GPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGSGSES 62
G PGTGKT V II ++ K++L + ++ A DN++ L N I +R+G
Sbjct 1074 GMPGTGKTTVIAEIIKIL--VSEGKRVLLTSYTHSAVDNILIKLRNTNISIMRLG----- 1126
Query 63 DLKEESIQDLARYRDLLRIRDKNSGEARSLRMLLVREAVRKYNVIIATCVGSGHEMF--D 120
+K + D +Y N +S L + + +V+ TC+G +F +
Sbjct 1127 -MKHKVHPDTQKYV-------PNYASVKSYNDYLSK--INSTSVVATTCLGINDILFTLN 1176
Query 121 DVTFERVIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLPPTI 164
+ F+ VI+DE +Q P L PL +G R +++GDH QLPP +
Sbjct 1177 EKDFDYVILDEASQISMPVALGPLRYGNR-FIMVGDHYQLPPLV 1219
> YKL017c
Length=683
Score = 64.3 bits (155), Expect = 9e-11, Method: Composition-based stats.
Identities = 57/206 (27%), Positives = 94/206 (45%), Gaps = 42/206 (20%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGSGS 60
+ GPPGTGKT +I NP ++IL SN++ D ++E L+ ++ + G
Sbjct 227 IHGPPGTGKTFTLIELIQQLLIKNPEERILICGPSNISVDTILERLTPLVPNNLLLRIGH 286
Query 61 ESDL--------------KEESIQDLARYRDLL----------RIRDKNSGEARSLRMLL 96
+ L K ++D+++ D L + R +N E + LR L
Sbjct 287 PARLLDSNKRHSLDILSKKNTIVKDISQEIDKLIQENKKLKNYKQRKENWNEIKLLRKDL 346
Query 97 -------VREAVRKYNVIIATCVGSGHEMF-----DDVTFE---RVIIDECAQSIEPSNL 141
+++ + + +++ T GS DD F+ +IIDE +Q++EP
Sbjct 347 KKREFKTIKDLIIQSRIVVTTLHGSSSRELCSLYRDDPNFQLFDTLIIDEVSQAMEPQCW 406
Query 142 IPL---GHGCRSVVLIGDHKQLPPTI 164
IPL + +VL GD+KQLPPTI
Sbjct 407 IPLIAHQNQFHKLVLAGDNKQLPPTI 432
> At4g15570
Length=525
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 50/158 (31%), Positives = 76/158 (48%), Gaps = 21/158 (13%)
Query 28 KILAVADSNVAADNLMEGLSNRGIRS----------VRVGSGSESDLKEESIQDLARYRD 77
++L A SN A D ++ L + G+R VR+G + + S+ L +
Sbjct 79 RVLVCAPSNSALDEIVLRLLSSGLRDENAQTYTPKIVRIGLKAHHSVASVSLDHLVAQKR 138
Query 78 LLRIRDKNSG----EARSLRMLLVREA--VRKYNV---IIATCVGSGHEMF--DDVTFER 126
I G + S+R ++ EA V +N + AT SG + + F+
Sbjct 139 GSAIDKPKQGTTGTDIDSIRTAILEEAAIVWGFNFRINVFATLSFSGSALLAKSNRGFDV 198
Query 127 VIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLPPTI 164
VIIDE AQ++EP+ LIPL C+ V L+GD KQLP T+
Sbjct 199 VIIDEAAQAVEPATLIPLATRCKQVFLVGDPKQLPATV 236
> At1g08840
Length=1296
Score = 59.7 bits (143), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 45/164 (27%), Positives = 76/164 (46%), Gaps = 18/164 (10%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGSGS 60
+ G PGTGKT + A + IL + +N A DNL+ L +GI +R+G
Sbjct 908 ILGMPGTGKTSTMVHAVKAL--LIRGSSILLASYTNSAVDNLLIKLKAQGIEFLRIGR-- 963
Query 61 ESDLKEESIQDLARYRDLLRIRDKNSGEARSLRMLLVREAVRKYNVIIATCVGSGHEMFD 120
+ + EE + ++ + D +++ + + V+ +TC+G +
Sbjct 964 DEAVHEEVRESCFSAMNMCSVED-------------IKKKLDQVKVVASTCLGINSPLLV 1010
Query 121 DVTFERVIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLPPTI 164
+ F+ IIDE Q P ++ PL + VL+GDH QLPP +
Sbjct 1011 NRRFDVCIIDEAGQIALPVSIGPLLFAS-TFVLVGDHYQLPPLV 1053
> YLR430w
Length=2231
Score = 58.2 bits (139), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 39/102 (38%), Positives = 59/102 (57%), Gaps = 9/102 (8%)
Query 69 IQDLARYRDLLRIRDKNSGEARSLRMLLVREA----VRKYNVIIATCVGSGHEMFDD--V 122
I +L R RD +R+KNS R+ R L R A + ++I +T GS H++ +
Sbjct 1526 INELGRDRD--EMREKNSVNYRN-RDLDRRNAQAHILAVSDIICSTLSGSAHDVLATMGI 1582
Query 123 TFERVIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLPPTI 164
F+ VIIDE Q E S++IPL +G + +++GD QLPPT+
Sbjct 1583 KFDTVIIDEACQCTELSSIIPLRYGGKRCIMVGDPNQLPPTV 1624
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 39/137 (28%), Positives = 57/137 (41%), Gaps = 40/137 (29%)
Query 1 VKGPPGTGKTHVACAIIDAW-----------------------QRINPTKKILAVADSNV 37
++GPPGTGKT II + +++ +KIL A SN
Sbjct 1355 IQGPPGTGKTKTILGIIGYFLSTKNASSSNVIKVPLEKNSSNTEQLLKKQKILICAPSNA 1414
Query 38 AADN----LMEGLSNR-----GIRSVRVGSGSESDLKEESIQDLARYRDLLRIRDKNSGE 88
A D L G+ ++ + VRVG SD+ +I+DL L + DK GE
Sbjct 1415 AVDEICLRLKSGVYDKQGHQFKPQLVRVG---RSDVVNVAIKDLT----LEELVDKRIGE 1467
Query 89 ARSLRMLLVREAVRKYN 105
R+ + E RK+N
Sbjct 1468 -RNYEIRTDPELERKFN 1483
> CE19303
Length=693
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 47/185 (25%), Positives = 85/185 (45%), Gaps = 23/185 (12%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGSGS 60
++GPPGTGKT V I+ Q + KK+L A ++VA N M+ + R I +
Sbjct 260 IQGPPGTGKTRVIAEIVH--QLMKKKKKVLVCAPTHVAVRNAMDATTGRMIEEMPEEKAE 317
Query 61 E-----SDLKEE--------SIQDLARYRDLLRIRD----KNSGEARSLRMLLVREAVRK 103
+ ++ K+E ++++ + + D K S E +R + R
Sbjct 318 QQLCLLANTKDEFQNHKSASKLEEIKKQLSTMSTSDPLYKKMSYEYYLIRNSIFRSIFYP 377
Query 104 YNVIIATCVGSGHEMFDDVTF--ERVIIDECAQSIEPSNLIPL--GHGCRSVVLIGDHKQ 159
+ +T S + + + + +I+DE AQ EP+ +P+ C+ ++L+GD KQ
Sbjct 378 KLAVFSTLGTSSIQKLPEYHWNADVMIVDEAAQCTEPATWVPVLTTPSCKKLILVGDQKQ 437
Query 160 LPPTI 164
LP +
Sbjct 438 LPAVV 442
> At1g05460
Length=1002
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 51/173 (29%), Positives = 77/173 (44%), Gaps = 18/173 (10%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEG-LSNRGIR-----SV 54
+ GPPGTGKT I ++L A SN AAD+++E L G+R
Sbjct 419 IHGPPGTGKTMTLVEAIVQLYTTQRNARVLVCAPSNSAADHILEKLLCLEGVRIKDNEIF 478
Query 55 RVGSGSESDLKEESIQDLARYRDLLRIRDKNSGEARSLRMLLVREAVRKYNVIIATCVGS 114
R+ + + S EE ++ R+ + K R LV +++ A V
Sbjct 479 RLNAATRS--YEEIKPEIIRFCFFDELIFKCPPLKALTRYKLVVSTYMSASLLNAEGVNR 536
Query 115 GHEMFDDVTFERVIIDECAQSIEPSNLIPLGHGCRS---VVLIGDHKQLPPTI 164
GH F +++DE Q+ EP N+I + + C + VVL GD +QL P I
Sbjct 537 GH-------FTHILLDEAGQASEPENMIAVSNLCLTETVVVLAGDPRQLGPVI 582
> At1g16800
Length=1939
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 34/115 (29%), Positives = 60/115 (52%), Gaps = 28/115 (24%)
Query 75 YRDLLRIRD---KNSGEARSLRMLLVREAVRKYNVIIATCVGSGHEMF------------ 119
Y+DL ++ K + E R+L+ L + +++ +++ T G G +++
Sbjct 1361 YKDLSAVQAQERKANYEMRTLKQKLRKSILKEAQIVVTTLSGCGGDLYSVCAESLAAHKF 1420
Query 120 ----DDVTFERVIIDECAQSIEPSNLIPL------GHGCRSVVLIGDHKQLPPTI 164
+D F+ V+IDE AQ++EP+ LIPL G C +++GD KQLP T+
Sbjct 1421 GSPSEDNLFDAVVIDEAAQALEPATLIPLQLLKSRGTKC---IMVGDPKQLPATV 1472
Score = 28.9 bits (63), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 11/17 (64%), Positives = 13/17 (76%), Gaps = 0/17 (0%)
Query 1 VKGPPGTGKTHVACAII 17
++GPPGTGKT AII
Sbjct 1129 IQGPPGTGKTRTIVAII 1145
> ECU05g1110
Length=563
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 55/178 (30%), Positives = 79/178 (44%), Gaps = 33/178 (18%)
Query 3 GPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRS--------- 53
GPPGTGKT II Q + +L SNV+ DN++E R +RS
Sbjct 197 GPPGTGKTRTVVEIIS--QLLAANSSVLVCGPSNVSVDNIIE----RFLRSRYFLCNQPS 250
Query 54 -VRVGSGSESDLKEESIQDLARYRDLLRIRDKNSGEARSLRMLLVREAVRKY-------- 104
R+GS ++ L ++ LA +K RS R L RE RK+
Sbjct 251 FYRLGSSTKG-LSHLNLDFLAESHTKFMKEEKGD---RSFRRDL-RERQRKFVEEKQANS 305
Query 105 NVIIATCVGSGHEMFDDVTFERVIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLPP 162
V+ AT S E F+ V++DE Q+ E + + + G R+ +L+GD QL P
Sbjct 306 PVVFATLFSSLKE---KRKFDWVLVDEACQASEAESFLAVVKG-RAFILVGDPMQLCP 359
> 7302834
Length=765
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 50/177 (28%), Positives = 72/177 (40%), Gaps = 20/177 (11%)
Query 3 GPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGSGSES 62
GPPGTGKT I + P +IL A SN A D + L ++R+
Sbjct 276 GPPGTGKTTTIVEAILQLRLQQPQSRILVTAGSNSACDTIALKLCEYIESNIRLQEHFAQ 335
Query 63 DLKEESIQDLAR----------YRDLLRIRDKNSGEARSLRMLLVREAVRKYNVIIATCV 112
E L R + + + KNS ++S+ + + KY +I+AT
Sbjct 336 QKLPEPDHQLIRVYSRSIYEKGFASVPSLLLKNSNCSKSIYDHIKASRIVKYGIIVATLC 395
Query 113 GSGHEMFDDV----TFERVIIDECAQSIEPSNLIPLGHGCRS-----VVLIGDHKQL 160
+ D + F + IDE S EP LI + G + V+L GDHKQL
Sbjct 396 TVARLVTDTLGRYNFFTHIFIDEAGASTEPEALIGI-MGIKQTADCHVILSGDHKQL 451
> Hs14251207_2
Length=776
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 61/177 (34%), Positives = 79/177 (44%), Gaps = 38/177 (21%)
Query 3 GPPGTGKTHVACAIIDAWQRIN---PTKKILAVADSNVAAD----NLMEGLSNRGIRSVR 55
GPPGTGKT II+A +++ P +IL A SN AAD L E + VR
Sbjct 335 GPPGTGKT---VTIIEAVLQVHFALPDSRILVCAPSNSAADLVCLRLHESKVLQPATMVR 391
Query 56 VGSGSESDLKEESIQDLARYRDLLRIRDKNSGEARSLRMLLVREAVRKYNVIIATCVGSG 115
V + +E I + Y RD GE + +A R + +II TC SG
Sbjct 392 VNATCR--FEEIVIDAVKPY-----CRD---GED-------IWKASR-FRIIITTCSSSG 433
Query 116 HEMFDDV-----TFERVIIDECAQSIEPSNLIPLGHGCR---SVVLIGDHKQLPPTI 164
+F + F V +DE Q+ EP LIPLG +VL GD QL P I
Sbjct 434 --LFYQIGVRVGHFTHVFVDEAGQASEPECLIPLGLMSDISGQIVLAGDPMQLGPVI 488
> Hs7662212
Length=828
Score = 51.2 bits (121), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 34/107 (31%), Positives = 55/107 (51%), Gaps = 12/107 (11%)
Query 66 EESIQDLARYRDLLRIRDKN-SGEARSLRMLLVREAVRKYNVIIATCVGSGHEMFDD--- 121
+E+I +++ R L + K G + + +++ E+ ++I T SG + +
Sbjct 264 DENISKVSKERQELASKIKEVQGRPQKTQSIIILES----HIICCTLSTSGGLLLESAFR 319
Query 122 ----VTFERVIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLPPTI 164
V F VI+DE QS E L PL H C ++L+GD KQLPPT+
Sbjct 320 GQGGVPFSCVIVDEAGQSCEIETLTPLIHRCNKLILVGDPKQLPPTV 366
> Hs14786304_1
Length=1839
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 34/89 (38%), Positives = 48/89 (53%), Gaps = 9/89 (10%)
Query 1 VKGPPGTGKTHVACAIIDA-------WQRINPTKKILAVADSNVAADNLMEGLSNRGIRS 53
++GPPGTGKT+V I+ A WQ IL V +N A D +EG+ N S
Sbjct 617 IQGPPGTGKTYVGLKIVQALLTNESVWQISLQKFPILVVCYTNHALDQFLEGIYNCQKTS 676
Query 54 -VRVGSGSESD-LKEESIQDLARYRDLLR 80
VRVG S S+ LK+ ++++L R+ R
Sbjct 677 IVRVGGRSNSEILKQFTLRELRNKREFRR 705
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 25/37 (67%), Gaps = 0/37 (0%)
Query 127 VIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLPPT 163
VI++E A+ +E + L C+ ++LIGDH+QL P+
Sbjct 1003 VIVEEAAEVLEAHTIATLSKACQHLILIGDHQQLRPS 1039
> Hs14211540
Length=1003
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 48/178 (26%), Positives = 73/178 (41%), Gaps = 25/178 (14%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGSGS 60
+ GPPGTGKT I + P ILA A SN AD L + L S+
Sbjct 522 IFGPPGTGKTVTLVEAIKQVVKHLPKAHILACAPSNSGADLLCQRLRVHLPSSIYRLLAP 581
Query 61 ESDLK--EESIQDLARYRDLLRIRDKNSGEARSLRMLLVREAVRKYNVIIATCVGSGH-- 116
D++ E I+ + D GE + ++ +++Y V+I T + +G
Sbjct 582 SRDIRMVPEDIKPCCNW-------DAKKGEY----VFPAKKKLQEYRVLITTLITAGRLV 630
Query 117 -EMFDDVTFERVIIDECAQSIEPSNLIPL---------GHGCRSVVLIGDHKQLPPTI 164
F F + IDE +EP +L+ + G +VL GD +QL P +
Sbjct 631 SAQFPIDHFTHIFIDEAGHCMEPESLVAIAGLMEVKETGDPGGQLVLAGDPRQLGPVL 688
> At2g19120
Length=1090
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/83 (39%), Positives = 44/83 (53%), Gaps = 7/83 (8%)
Query 85 NSGEAR-SLRMLLVREAVRKYNVIIATCVGSGHEMFDDVT--FERVIIDECAQSIEPSNL 141
N EAR SL EA ++ T SG ++F +T F+ V+IDE AQ+ E L
Sbjct 737 NLEEARASLEASFANEA----EIVFTTVSSSGRKLFSRLTHGFDMVVIDEAAQASEVGVL 792
Query 142 IPLGHGCRSVVLIGDHKQLPPTI 164
PL G VL+GD +QLP T+
Sbjct 793 PPLALGAARCVLVGDPQQLPATV 815
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 10/18 (55%), Positives = 14/18 (77%), Gaps = 0/18 (0%)
Query 1 VKGPPGTGKTHVACAIID 18
V+GPPGTGKTH +++
Sbjct 500 VQGPPGTGKTHTVWGMLN 517
> At4g30100
Length=1311
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/83 (39%), Positives = 44/83 (53%), Gaps = 7/83 (8%)
Query 85 NSGEAR-SLRMLLVREAVRKYNVIIATCVGSGHEMFDDVT--FERVIIDECAQSIEPSNL 141
N EAR SL EA ++ T SG ++F +T F+ V+IDE AQ+ E L
Sbjct 900 NLEEARASLEASFANEA----EIVFTTVSSSGRKLFSRLTHGFDMVVIDEAAQASEVGVL 955
Query 142 IPLGHGCRSVVLIGDHKQLPPTI 164
PL G VL+GD +QLP T+
Sbjct 956 PPLALGAARCVLVGDPQQLPATV 978
Score = 30.0 bits (66), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 10/18 (55%), Positives = 14/18 (77%), Gaps = 0/18 (0%)
Query 1 VKGPPGTGKTHVACAIID 18
V+GPPGTGKTH +++
Sbjct 664 VQGPPGTGKTHTVWGMLN 681
> YER176w
Length=1121
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 54/185 (29%), Positives = 82/185 (44%), Gaps = 26/185 (14%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEG-LSNR-GIRSVRVGS 58
++GPPGTGKT II IL VA SN+A DN+ E + NR I+ +R+ S
Sbjct 668 LQGPPGTGKTSTIEEIIIQVIERFHAFPILCVAASNIAIDNIAEKIMENRPQIKILRILS 727
Query 59 -------GSESDLKEESIQDLARYRDLLRIRDKNSGEARSLRML--------------LV 97
+ L E + ++ Y++L + + R M+ +
Sbjct 728 KKKEQQYSDDHPLGEICLHNIV-YKNLSPDMQVVANKTRRGEMISKSEDTKFYKEKNRVT 786
Query 98 REAVRKYNVIIATCVGSGHEMFDDVT-FERVIIDECAQSIEPSNLIPLG-HGCRSVVLIG 155
+ V + +I T + +G + VI+DE QS E S L+PL G R+ V +G
Sbjct 787 NKVVSQSQIIFTTNIAAGGRELKVIKECPVVIMDEATQSSEASTLVPLSLPGIRNFVFVG 846
Query 156 DHKQL 160
D KQL
Sbjct 847 DEKQL 851
> ECU09g1800
Length=934
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 34/162 (20%), Positives = 72/162 (44%), Gaps = 33/162 (20%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGSGS 60
+ G PGTGK+ + C +I + KK+L + +N+A N+ + L+ +R+ +
Sbjct 643 IHGMPGTGKSTLICLLIKILAYLK--KKVLLICYTNLALANITKKLN-----GIRIYAAG 695
Query 61 ESDLKEESIQDLARYRDLLRIRDKNSGEARSLRMLLVREAVRKYNVIIATCVGSGHEMFD 120
+ ++ +++++ + D + +++ TC ++
Sbjct 696 KEEVSFKTVEEAGLFFD-------------------------RIELVVGTCFSFADPIYV 730
Query 121 DVTFERVIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLPP 162
+ F+ +IDE +Q LIP+ + V++GDH QL P
Sbjct 731 NRQFDFCVIDEGSQMHLLLTLIPVSIS-KKFVIVGDHLQLKP 771
> SPBC16D10.04c
Length=1398
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 42/166 (25%), Positives = 71/166 (42%), Gaps = 21/166 (12%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGSGS 60
+ G PGTGKT ++I + + + +++A DN++ L VR+GS
Sbjct 953 ILGMPGTGKTTTISSLIRSLLAKKKKILLTSF--THLAVDNILIKLKGCDSTIVRLGSPH 1010
Query 61 ESD--LKEESIQDLARYRDLLRIRDKNSGEARSLRMLLVREAVRKYNVIIATCVGSGHEM 118
+ +KE + + + DL ++ ++ + +G H +
Sbjct 1011 KIHPLVKEFCLTEGTTFDDLASLK----------------HFYEDPQIVACSSLGVYHSI 1054
Query 119 FDDVTFERVIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLPPTI 164
F+ F+ IIDE +Q P L PL VL+GDH QLPP +
Sbjct 1055 FNKRKFDYCIIDEASQIPLPICLGPL-QLAEKFVLVGDHYQLPPLV 1099
> Hs21703358_3
Length=498
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 53/227 (23%), Positives = 85/227 (37%), Gaps = 65/227 (28%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKK----------------ILAVADSNVAADNLME 44
++GPPGTGKT V I+ + + N + IL SN + D L
Sbjct 21 IQGPPGTGKTIVGLHIVFWFHKSNQEQVQPGGPPRGEKRLGGPCILYCGPSNKSVDVLAG 80
Query 45 GLSNR-GIRSVRVGSGSESDLKEESIQDLARYRDLLRIRDKNSGEARSLRMLLVREAVR- 102
L R ++ +RV S +++ E + + R LLR + +SLR + + +R
Sbjct 81 LLLRRMELKPLRVYS-EQAEASEFPVPRVGS-RKLLRKSPREGRPNQSLRSITLHHRIRQ 138
Query 103 -------------------------------------------KYNVIIATCVGSGHEMF 119
++ VI+ TC +
Sbjct 139 APNPYSSEIKAFDTRLQRGELFSREDLVWYKKVLWEARKFELDRHEVILCTCSCAASASL 198
Query 120 DDVTFERVIIDECAQSIEPSNLIPLGH--GCRSVVLIGDHKQLPPTI 164
+ ++++DE + EP LIPL VVL+GDHKQL P +
Sbjct 199 KILDVRQILVDEAGMATEPETLIPLVQFPQAEKVVLLGDHKQLRPVV 245
> Hs14769950_3
Length=498
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 53/227 (23%), Positives = 85/227 (37%), Gaps = 65/227 (28%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKK----------------ILAVADSNVAADNLME 44
++GPPGTGKT V I+ + + N + IL SN + D L
Sbjct 21 IQGPPGTGKTIVGLHIVFWFHKSNQEQVQPGGPPRGEKRLGGPCILYCGPSNKSVDVLAG 80
Query 45 GLSNR-GIRSVRVGSGSESDLKEESIQDLARYRDLLRIRDKNSGEARSLRMLLVREAVR- 102
L R ++ +RV S +++ E + + R LLR + +SLR + + +R
Sbjct 81 LLLRRMELKPLRVYS-EQAEASEFPVPRVGS-RKLLRKSPREGRPNQSLRSITLHHRIRQ 138
Query 103 -------------------------------------------KYNVIIATCVGSGHEMF 119
++ VI+ TC +
Sbjct 139 APNPYSSEIKAFDTRLQRGELFSREDLVWYKKVLWEARKFELDRHEVILCTCSCAASASL 198
Query 120 DDVTFERVIIDECAQSIEPSNLIPLGH--GCRSVVLIGDHKQLPPTI 164
+ ++++DE + EP LIPL VVL+GDHKQL P +
Sbjct 199 KILDVRQILVDEAGMATEPETLIPLVQFPQAEKVVLLGDHKQLRPVV 245
> 7299528
Length=1170
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/78 (38%), Positives = 41/78 (52%), Gaps = 6/78 (7%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRS---VRVG 57
V GPPGTGKT VA II +P ++ L V SN A + L E + I +R+G
Sbjct 518 VVGPPGTGKTDVAVQIISNIYHNHPNQRTLIVTHSNQALNQLFEKIMALDIDERHLLRLG 577
Query 58 SGSESDLKEESIQDLARY 75
G E+ E+ +D +RY
Sbjct 578 HGEEA---LETEKDYSRY 592
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 31/108 (28%), Positives = 52/108 (48%), Gaps = 20/108 (18%)
Query 69 IQDLARYRDLLRIRDKNSGEARSLRMLLVREAVRKYNVIIATCVGSGHEMFDDVT----F 124
+L +R +R +G RS + LLV+EA +I TC + + + V +
Sbjct 705 FNELEEFRAFELLR---TGLDRS-KYLLVKEA----KIIAMTCTHAALKRKELVNLGFRY 756
Query 125 ERVIIDECAQSIEPSNLIPLG--------HGCRSVVLIGDHKQLPPTI 164
+ ++++E AQ +E IPL + + ++IGDH QLPP I
Sbjct 757 DNILMEESAQILEIETFIPLLLQNPLDGLNRLKRWIMIGDHHQLPPVI 804
> CE23725
Length=1069
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 49/167 (29%), Positives = 80/167 (47%), Gaps = 16/167 (9%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNR--GIRSVRVGS 58
V+G PG+GKT + +I N KK+L A ++ A DN++ L+ + +R+GS
Sbjct 670 VEGLPGSGKTTLISVLIQCLVATN--KKVLLAAFTHSAVDNILTKLTKEVAAEKILRLGS 727
Query 59 GSESDLKEESIQDLARYRDLLRIRDKNSGEARSLRMLLVREAVRKYNVIIATCVGSGHEM 118
S SI+D + + ++ K E VR+ ++ ++ TC E+
Sbjct 728 SS-------SIKDDIKK---MTLKAKLENETSEEYYAAVRKVMKTTPIVACTCHHVPREL 777
Query 119 -FDDVTFERVIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLPPTI 164
F F+ VI+DE + +EP L L + VL+GD KQL P +
Sbjct 778 LFSYRHFDVVIVDEASMVLEPLLLPVLATSNK-FVLVGDCKQLTPLV 823
> At1g65780
Length=1065
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 42/166 (25%), Positives = 77/166 (46%), Gaps = 22/166 (13%)
Query 10 THVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGSGSESDLKEESI 69
TH+ A++ + + I V D + A +++G++ G++SV + +G SD S
Sbjct 451 THLPTALLSSQAATRMYEAIDLVRDVTILA--ILDGVTGEGVKSVLIPNGEGSD--RFSS 506
Query 70 QDLARYRDLLRIRDKNSGEARSLRMLLVREAVRKYNVIIATCVGSGHEMFDDVT------ 123
Q + D L++ RS+ + AV ++I C+G +F +
Sbjct 507 QHVTVEDDYLKL-------LRSIPEIFPLPAVSDRHLIKELCLGHACLLFSTASCSARLY 559
Query 124 ----FERVIIDECAQSIEPSNLIPLG-HGCRSVVLIGDHKQLPPTI 164
+ ++IDE AQ E + IP+ G R ++L+GD +QLP +
Sbjct 560 TGTPIQLLVIDEAAQLKECESSIPMQLPGLRHLILVGDERQLPAMV 605
> CE02219
Length=1105
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 49/167 (29%), Positives = 80/167 (47%), Gaps = 16/167 (9%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNR--GIRSVRVGS 58
V+G PG+GKT + +I N KK+L A ++ A DN++ L+ + +R+GS
Sbjct 706 VEGLPGSGKTTLISVLIQCLVATN--KKVLLAAFTHSAVDNILTKLTKEVAAEKILRLGS 763
Query 59 GSESDLKEESIQDLARYRDLLRIRDKNSGEARSLRMLLVREAVRKYNVIIATCVGSGHEM 118
S SI+D + + ++ K E VR+ ++ ++ TC E+
Sbjct 764 SS-------SIKDDIKK---MTLKAKLENETSEEYYAAVRKVMKTTPIVACTCHHVPREL 813
Query 119 -FDDVTFERVIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLPPTI 164
F F+ VI+DE + +EP L L + VL+GD KQL P +
Sbjct 814 LFSYRHFDVVIVDEASMVLEPLLLPVLATSNK-FVLVGDCKQLTPLV 859
> 7303245
Length=1194
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 51/185 (27%), Positives = 79/185 (42%), Gaps = 38/185 (20%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGSGS 60
V GPPGTGKT I P IL +A SN A D ++ R +R++ S
Sbjct 496 VFGPPGTGKTSTIVEAIYQLYINRPETHILVLAGSNTACDE----VALRLLRAIAKAPQS 551
Query 61 ES-----------DLKEESIQDLARYRDLLRIRDKNSGEARSLRMLLVREAVRKYNVIIA 109
+ D + ++I DL LL + ++ +AV +Y ++I
Sbjct 552 QPRPLTRIFSANCDRRIDNIDDL-----LLEYSN-----MYTVHFYPAVQAVHQYRIVIC 601
Query 110 TC-----VGSGHEMFDDVTFERVIIDECAQSIEPSNLIPLGHGCR-----SVVLIGDHKQ 159
T + +G ++V + V +DE A S E L +G C +++L GDHKQ
Sbjct 602 TLSLAGKLSTGGFAANNV-YTHVFVDEAAASTEAEVL--MGITCTLSPTLNLILSGDHKQ 658
Query 160 LPPTI 164
L P +
Sbjct 659 LGPVL 663
> Hs14750549
Length=1485
Score = 44.7 bits (104), Expect = 9e-05, Method: Composition-based stats.
Identities = 30/78 (38%), Positives = 40/78 (51%), Gaps = 6/78 (7%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRS---VRVG 57
V GPPGTGKT VA II P ++ L V SN A + L E + I +R+G
Sbjct 821 VVGPPGTGKTDVAVQIISNIYHNFPEQRTLIVTHSNQALNQLFEKIMALDIDERHLLRLG 880
Query 58 SGSESDLKEESIQDLARY 75
G E + E+ +D +RY
Sbjct 881 HGEE---ELETEKDFSRY 895
Score = 42.0 bits (97), Expect = 6e-04, Method: Composition-based stats.
Identities = 35/103 (33%), Positives = 51/103 (49%), Gaps = 22/103 (21%)
Query 74 RYRDLLRIRDKNSGEARSLRMLLVREAVRKYNVIIATCVGSGHEMFDDVT----FERVII 129
R +LLR SG RS + LLV+EA +I TC + + D V ++ +++
Sbjct 1012 RASELLR-----SGLDRS-KYLLVKEA----KIIAMTCTHAALKRHDLVKLGFKYDNILM 1061
Query 130 DECAQSIEPSNLIPL-------GHG-CRSVVLIGDHKQLPPTI 164
+E AQ +E IPL G + ++IGDH QLPP I
Sbjct 1062 EEAAQILEIETFIPLLLQNPQDGFSRLKRWIMIGDHHQLPPVI 1104
> At2g38770
Length=1444
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 31/98 (31%), Positives = 47/98 (47%), Gaps = 12/98 (12%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRS---VRVG 57
V GPPGTGKT A I++ P+++ L + SN A ++L E + R + + +R+G
Sbjct 885 VVGPPGTGKTDTAVQILNVLYHNCPSQRTLIITHSNQALNDLFEKIMERDVPARYLLRLG 944
Query 58 SGSESDLKEESIQDLARYRDLLRIRDKNSGEARSLRML 95
G Q+LA D R N+ R L +L
Sbjct 945 QGE---------QELATDLDFSRQGRVNAMLVRRLELL 973
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 28/49 (57%), Gaps = 8/49 (16%)
Query 124 FERVIIDECAQSIEPSNLIPL-------GHG-CRSVVLIGDHKQLPPTI 164
++ ++++E AQ +E IP+ GH + +LIGDH QLPP +
Sbjct 1117 YDNLLMEESAQILEIETFIPMLLQRQEDGHARLKRCILIGDHHQLPPVV 1165
> 7292369_2
Length=754
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 48/174 (27%), Positives = 70/174 (40%), Gaps = 17/174 (9%)
Query 3 GPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRG-------IRSVR 55
GPPG+GKT + R P +IL SN +AD + + L + IR V
Sbjct 289 GPPGSGKTMTLIETLLQLVRNLPGARILVGTPSNSSADLVTKRLIDSKALLQGDFIRLVS 348
Query 56 VGSGSESDLKEESIQDLARYRDLLRI---RDKNSGEARSLRMLLVREAVRKYNVIIATCV 112
E DL I D+ + DK L++ + + + + I+TC
Sbjct 349 YNQ-VEKDLIPPEIMSYCATSDVGAVGSCEDKMMVTESGLKLRCQAKFIGTHRITISTCT 407
Query 113 GSGHEM---FDDVTFERVIIDECAQSIEPSNLIP---LGHGCRSVVLIGDHKQL 160
G+ + F F V+ DE Q EP ++P L VVL GD +QL
Sbjct 408 TLGNFLQLGFPAGHFTHVLFDEAGQCTEPETMVPIVMLTKKRSQVVLSGDPRQL 461
> CE24674
Length=1212
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 29/78 (37%), Positives = 40/78 (51%), Gaps = 6/78 (7%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRS---VRVG 57
V GPPGTGKT VA II P ++ L V SN A + L E + + +R+G
Sbjct 451 VVGPPGTGKTDVAVQIISNIYHNWPNQRTLIVTHSNQALNQLFEKIIALDVDERHLLRMG 510
Query 58 SGSESDLKEESIQDLARY 75
G E+ E+ +D +RY
Sbjct 511 HGEEA---LETEKDFSRY 525
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 26/97 (26%), Positives = 45/97 (46%), Gaps = 17/97 (17%)
Query 85 NSGEARSLRMLLVR-----EAVRKYNVIIATCVGSGHEMFDDVT----FERVIIDECAQS 135
N+ R+ ML + V++ +I TC + + V ++ ++++E AQ
Sbjct 690 NTNRRRTRSMLFLDLCSFLSLVKEAKIIAMTCTHAALRRNELVKLGFRYDNIVMEEAAQI 749
Query 136 IEPSNLIPL-------GHG-CRSVVLIGDHKQLPPTI 164
+E IPL GH + ++IGDH QLPP +
Sbjct 750 LEVETFIPLLLQNPQDGHNRLKRWIMIGDHHQLPPVV 786
> CE08664
Length=633
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 46/198 (23%), Positives = 85/198 (42%), Gaps = 47/198 (23%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVGSGS 60
++GPPGTGKT ++ + I K+++ +A + A N+ ++ + ++ + +
Sbjct 95 IQGPPGTGKTFTLTLLL--CRLIQQKKQVVVLAPTREALANI-RMMTKKTLKRMGI---- 147
Query 61 ESDLKEESIQDLARYRDLLRIRDK---NSGEARSLRMLL------------VREAV--RK 103
+ E ++ D YRD++ D+ + E R LR +R+++ R
Sbjct 148 --KVHEHALMDTNEYRDVINKSDRALMAAEEVRDLRKAFDNGEITENVLDEMRQSIINRV 205
Query 104 YNVIIATCVGS-----------------GHEMFDDVTFERVIIDECAQSIEPSNLIPLGH 146
N + A +G+ H+ FD IIDE AQ +E P +
Sbjct 206 RNEVGAEVIGNVRVAFATIGASFVDFVMKHKKFDPCL---CIIDEAAQVMEAQTW-PAVY 261
Query 147 GCRSVVLIGDHKQLPPTI 164
+ +V+ GD KQLP +
Sbjct 262 KMKRIVMAGDPKQLPALV 279
> 7295157
Length=1417
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 34/59 (57%), Gaps = 1/59 (1%)
Query 107 IIATCVGSGHEMFDDVTFERV-IIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLPPTI 164
II T + S ++ + V F + I+DE Q EP L+P+ G +VL+GD +QLP +
Sbjct 1108 IICTTLSSCVKLANYVDFFDICIVDEATQCTEPWTLLPMRFGLTHMVLVGDMQQLPAVV 1166
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 25/47 (53%), Gaps = 4/47 (8%)
Query 1 VKGPPGTGKTHVACAIIDAWQRINPTK----KILAVADSNVAADNLM 43
++GPPGTGK+ V + N K KIL A SN A D+++
Sbjct 939 IQGPPGTGKSRVISELCLQTLYGNAAKTLDRKILICAHSNTAVDHIV 985
> 7301121
Length=903
Score = 42.0 bits (97), Expect = 6e-04, Method: Composition-based stats.
Identities = 46/212 (21%), Positives = 80/212 (37%), Gaps = 52/212 (24%)
Query 1 VKGPPGTGKTHVACAIIDAW---QRINPTKKILAVADSNVAADNLMEGLSNRGIRSVRVG 57
++GPPGTGKTH++ ++++ + T I+ + +N + D + +S +R G
Sbjct 469 IQGPPGTGKTHLSVQLVNSLIQNAKALGTGPIIVLTYTNNSLDKFLVKISRYTQEILRFG 528
Query 58 SGSES----------------------------------------------DLKEESIQD 71
+ S D EES QD
Sbjct 529 NQSRDPQISKFNLSTTIKPELVPPRLKRIWWLVNCEYKEKFRNLQGLYANFDGSEESYQD 588
Query 72 -LARYRDLLRIRDKNSGEARSLRMLLVREAVRKYNVIIATCVGSGHEMFDDVTFERVIID 130
LA L ++ ++ + L RE + + TC + +F + + V+ +
Sbjct 589 TLAAQEKLNQVAERIETLRMVFQFFLARE--KDLLAMTTTCAARHNFLFRLLQSKCVLFE 646
Query 131 ECAQSIEPSNLIPLGHGCRSVVLIGDHKQLPP 162
E A+ E + L V+L+GDHKQL P
Sbjct 647 EAAEIQEAHIVACLTPHTEHVILVGDHKQLQP 678
> CE06677
Length=2219
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 27/51 (52%), Gaps = 10/51 (19%)
Query 1 VKGPPGTGKTHVACAIIDA-------WQRINPTKKILAVADSNVAADNLME 44
++GPPGTGKTH+ I+ W+ P IL V +N DNL+E
Sbjct 828 IQGPPGTGKTHIGVQIVKTILQNRSYWKITEP---ILVVCFTNSGLDNLLE 875
Score = 28.9 bits (63), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 127 VIIDECAQSIEPSNLIPLGHGCRSVVLIGDHKQLPP 162
+I++E A+ +E + + VV+IGDHKQL P
Sbjct 1233 LIVEEAAEVLEAHIISAMISTVEHVVMIGDHKQLRP 1268
> Hs7661884
Length=1942
Score = 38.9 bits (89), Expect = 0.005, Method: Composition-based stats.
Identities = 24/81 (29%), Positives = 43/81 (53%), Gaps = 5/81 (6%)
Query 89 ARSLRMLLVREAVRKYNVIIATCVGSGHEMFDDVT---FERVIIDECAQSIEPSNLIPLG 145
A S + +E + K+ V++ T S + D+ F +++DE AQ++E ++PL
Sbjct 750 AHSTFQMPQKEDILKHRVVVVTLNTSQYLCQLDLEPGFFTHILLDEAAQAMECETIMPLA 809
Query 146 HGCRS--VVLIGDHKQLPPTI 164
++ +VL GDH QL P +
Sbjct 810 LATQNTRIVLAGDHMQLSPFV 830
Lambda K H
0.319 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2317343104
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40