bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1494_orf1
Length=146
Score E
Sequences producing significant alignments: (Bits) Value
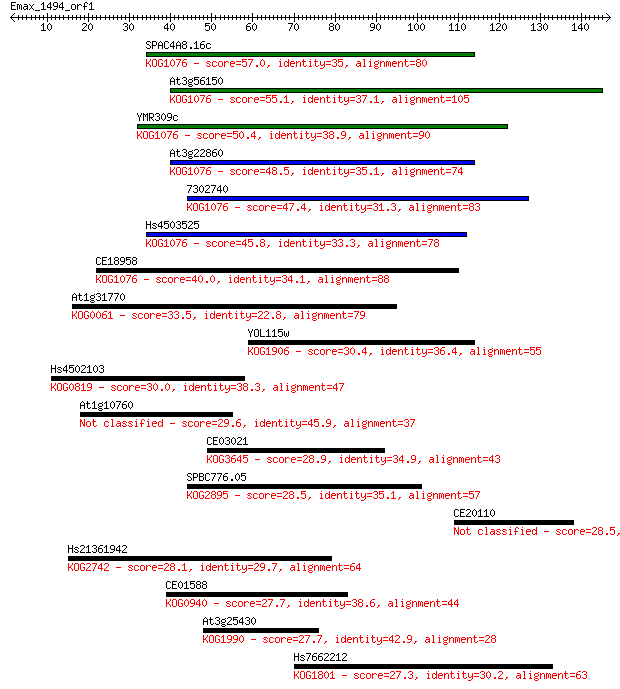
SPAC4A8.16c 57.0 1e-08
At3g56150 55.1 4e-08
YMR309c 50.4 1e-06
At3g22860 48.5 4e-06
7302740 47.4 9e-06
Hs4503525 45.8 3e-05
CE18958 40.0 0.001
At1g31770 33.5 0.14
YOL115w 30.4 1.4
Hs4502103 30.0 1.6
At1g10760 29.6 2.2
CE03021 28.9 3.2
SPBC776.05 28.5 4.2
CE20110 28.5 4.6
Hs21361942 28.1 5.8
CE01588 27.7 7.7
At3g25430 27.7 7.7
Hs7662212 27.3 9.7
> SPAC4A8.16c
Length=639
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 48/80 (60%), Gaps = 1/80 (1%)
Query 34 QISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKT 93
Q+ S++ SF+ERLDD+ +SLQ ID H+ EY +RL + LL R+ +L+ G
Sbjct 167 QVQGSVV-SFLERLDDEFTRSLQMIDPHTPEYIDRLKDETSLYTLLVRSQGYLERIGVVE 225
Query 94 QAATVALKVNEHMHYKPDNI 113
A + ++ + ++YKP+ +
Sbjct 226 NTARLIMRRLDRVYYKPEQV 245
> At3g56150
Length=900
Score = 55.1 bits (131), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 39/105 (37%), Positives = 55/105 (52%), Gaps = 0/105 (0%)
Query 40 LTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKTQAATVA 99
L +F+ER+D + KSLQ ID H+ EY ERL LAL + + G AA VA
Sbjct 356 LVAFLERVDTEFFKSLQCIDPHTREYVERLRDEPMFLALAQNIQDYFERMGDFKAAAKVA 415
Query 100 LKVNEHMHYKPDNIAIKMWDVLRKELPEELCKDLPGENIPPTAFI 144
L+ E ++YKP + M + EE ++ E+ PPT+FI
Sbjct 416 LRRVEAIYYKPQEVYDAMRKLAELVEEEEETEEAKEESGPPTSFI 460
> YMR309c
Length=812
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 35/99 (35%), Positives = 53/99 (53%), Gaps = 10/99 (10%)
Query 32 IQQISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRTFCFL----- 86
+++I SI SFVERLDD+ MKSL +ID HS +Y RL I L+ RT +
Sbjct 362 VKRILGSIF-SFVERLDDEFMKSLLNIDPHSSDYLIRLRDEQSIYNLILRTQLYFEATLK 420
Query 87 DERGFKTQAATVALKVNEHMHYKPDNIAIKM----WDVL 121
DE + +K +H++YK +N+ M W+++
Sbjct 421 DEHDLERALTRPFVKRLDHIYYKSENLIKIMETAAWNII 459
> At3g22860
Length=800
Score = 48.5 bits (114), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 42/74 (56%), Gaps = 0/74 (0%)
Query 40 LTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKTQAATVA 99
L +F+E+++ + KSLQ ID H+++Y ERL LAL +L+ G A+ VA
Sbjct 313 LVAFLEKIETEFFKSLQCIDPHTNDYVERLKDEPMFLALAQSIQDYLERTGDSKAASKVA 372
Query 100 LKVNEHMHYKPDNI 113
+ E ++YKP +
Sbjct 373 FILVESIYYKPQEV 386
> 7302740
Length=910
Score = 47.4 bits (111), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 44/83 (53%), Gaps = 3/83 (3%)
Query 44 VERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKTQAATVALKVN 103
VERLDD+ +K L+ D HS++Y RL ++++ + + + G + + L+
Sbjct 425 VERLDDEFVKLLKECDPHSNDYVSRLKDEVNVVKTIELVLQYFERSGTNNERCRIYLRKI 484
Query 104 EHMHYKPDNIAIKMWDVLRKELP 126
EH++YK D +K R ELP
Sbjct 485 EHLYYKFDPEVLKK---KRGELP 504
> Hs4503525
Length=913
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 42/78 (53%), Gaps = 1/78 (1%)
Query 34 QISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKT 93
++ ILT VER+D++ K +Q+ D HS EY E L + A++ R +L+E+G
Sbjct 439 RVRGCILT-LVERMDEEFTKIMQNTDPHSQEYVEHLKDEAQVCAIIERVQRYLEEKGTTE 497
Query 94 QAATVALKVNEHMHYKPD 111
+ + L H +YK D
Sbjct 498 EVCRIYLLRILHTYYKFD 515
> CE18958
Length=898
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 51/93 (54%), Gaps = 8/93 (8%)
Query 22 EAITETTEEEIQQISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWR 81
E + + T+E +I SIL + V+RLD +L K LQ+ D HS++Y E+L D+ +L+ +
Sbjct 404 ENLKDDTQE--YRIQGSILIA-VQRLDGELAKILQNADCHSNDYIEKLKAEKDMCSLIEK 460
Query 82 TFCFLDERG-----FKTQAATVALKVNEHMHYK 109
+++ R K + V + EH +YK
Sbjct 461 AEKYVELRNDSGIFDKHEVCKVYMMRIEHAYYK 493
> At1g31770
Length=646
Score = 33.5 bits (75), Expect = 0.14, Method: Composition-based stats.
Identities = 18/82 (21%), Positives = 40/82 (48%), Gaps = 3/82 (3%)
Query 16 AQGVTTEAITETTEEEIQQISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSI-- 73
A G+ + ET+E+E + + ++++++ + + L L + + HS EY + K++
Sbjct 314 ANGIPPDTQKETSEQEQKTVKETLVSAYEKNISTKLKAELCNAESHSYEYTKAAAKNLKS 373
Query 74 -DILALLWRTFCFLDERGFKTQ 94
W F L +RG + +
Sbjct 374 EQWCTTWWYQFTVLLQRGVRER 395
> YOL115w
Length=584
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 30/57 (52%), Gaps = 5/57 (8%)
Query 59 DVHSDEYKERLGKSI--DILALLWRTFCFLDERGFKTQAATVALKVNEHMHYKPDNI 113
++HS +K+RLGKSI +++ + F DERG A + NE+ H K I
Sbjct 460 ELHSATFKDRLGKSILGNVIKYRGKARDFKDERGLVLNKAIIE---NENYHKKRSRI 513
> Hs4502103
Length=338
Score = 30.0 bits (66), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 23/47 (48%), Gaps = 0/47 (0%)
Query 11 LEATEAQGVTTEAITETTEEEIQQISASILTSFVERLDDDLMKSLQS 57
L A QGV AI + ++ I +F ER DLMKSLQ+
Sbjct 44 LRAITGQGVDRSAIVDVLTNRSREQRQLISRNFQERTQQDLMKSLQA 90
> At1g10760
Length=1399
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 17/37 (45%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 18 GVTTEAITETTEEEIQQISASILTSFVERLDDDLMKS 54
GV A++ TEE I+ SA+ L+S V RLD L K+
Sbjct 901 GVDQSAVSIFTEEIIRAGSAAALSSLVNRLDPVLRKT 937
> CE03021
Length=814
Score = 28.9 bits (63), Expect = 3.2, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 23/43 (53%), Gaps = 6/43 (13%)
Query 49 DDLMKSLQSIDVHSDEYKERLGKSIDILALLWRTFCFLDERGF 91
D L +L +D+HS G + +L +TFCFLD +G+
Sbjct 136 DLLSNTLVEMDIHSK------GFVMTTTKILLKTFCFLDFKGY 172
> SPBC776.05
Length=404
Score = 28.5 bits (62), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 28/60 (46%), Gaps = 3/60 (5%)
Query 44 VERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLW---RTFCFLDERGFKTQAATVAL 100
V R + L + I+ +D+ KERLGKSID W + F D+ F +T L
Sbjct 65 VSRQKNKLQPIHKQINYETDKLKERLGKSIDKFQEQWNSGKVVRFRDKLSFALGVSTCIL 124
> CE20110
Length=194
Score = 28.5 bits (62), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 12/34 (35%), Positives = 21/34 (61%), Gaps = 5/34 (14%)
Query 109 KPDNIAIKMWD-----VLRKELPEELCKDLPGEN 137
KP KMW +++ +LP+ELCK++ G++
Sbjct 124 KPSPECYKMWGPFDGFIIKDKLPDELCKNMVGKD 157
> Hs21361942
Length=385
Score = 28.1 bits (61), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 32/64 (50%), Gaps = 1/64 (1%)
Query 15 EAQGVTTEAITETTEEEIQQISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSID 74
A+G T EA+ TEEE +Q++ + +F DD++ Q +D+ L + I
Sbjct 24 RAEGFTVEALWGKTEEEAKQLAEEMNIAFYTSRTDDILLH-QDVDLVCISIPPPLTRQIS 82
Query 75 ILAL 78
+ AL
Sbjct 83 VKAL 86
> CE01588
Length=889
Score = 27.7 bits (60), Expect = 7.7, Method: Composition-based stats.
Identities = 17/44 (38%), Positives = 27/44 (61%), Gaps = 5/44 (11%)
Query 39 ILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRT 82
+LTSF + L+D+L+ SL+S D+ K L S+++ WRT
Sbjct 742 LLTSFNQILNDNLLNSLESSDL-----KRILSGSLELDLNDWRT 780
> At3g25430
Length=618
Score = 27.7 bits (60), Expect = 7.7, Method: Composition-based stats.
Identities = 12/28 (42%), Positives = 20/28 (71%), Gaps = 0/28 (0%)
Query 48 DDDLMKSLQSIDVHSDEYKERLGKSIDI 75
DDDL+ SL+ + + S++Y RL +ID+
Sbjct 216 DDDLVSSLRRLVLGSEQYGSRLCLTIDV 243
> Hs7662212
Length=828
Score = 27.3 bits (59), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 32/65 (49%), Gaps = 4/65 (6%)
Query 70 GKSIDILALLWRTFCFLDERGFKTQAATVALKVNEHMHYKPDNIAIKMWDVLRKELPE-- 127
GKS I+ LL+R +G + + +K N + P N A+ ++++K + E
Sbjct 119 GKSKTIVGLLYRLLTENQRKGHSDENSNAKIKQNRVLVCAPSNAAVD--ELMKKIILEFK 176
Query 128 ELCKD 132
E CKD
Sbjct 177 EKCKD 181
Lambda K H
0.315 0.132 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1748847648
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40