bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1386_orf1
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
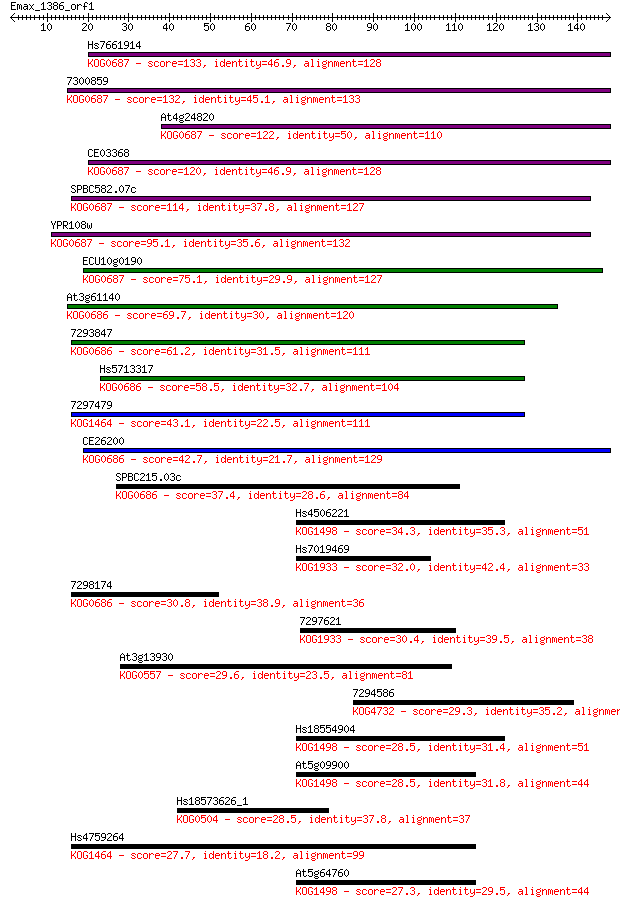
Hs7661914 133 2e-31
7300859 132 2e-31
At4g24820 122 3e-28
CE03368 120 8e-28
SPBC582.07c 114 8e-26
YPR108w 95.1 4e-20
ECU10g0190 75.1 4e-14
At3g61140 69.7 2e-12
7293847 61.2 7e-10
Hs5713317 58.5 4e-09
7297479 43.1 2e-04
CE26200 42.7 3e-04
SPBC215.03c 37.4 0.012
Hs4506221 34.3 0.091
Hs7019469 32.0 0.39
7298174 30.8 1.1
7297621 30.4 1.2
At3g13930 29.6 2.4
7294586 29.3 3.2
Hs18554904 28.5 4.7
At5g09900 28.5 4.7
Hs18573626_1 28.5 4.8
Hs4759264 27.7 8.3
At5g64760 27.3 9.4
> Hs7661914
Length=389
Score = 133 bits (334), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 60/128 (46%), Positives = 88/128 (68%), Gaps = 0/128 (0%)
Query 20 LNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLINM 79
L + Y +Y F Q L + + +K D +PHY Y++R +R+ AY Q L+ Y+S+TL M
Sbjct 260 LFSLYECRYSVFFQSLAVVEQEMKKDWLFAPHYRYYVREMRIHAYSQLLESYRSLTLGYM 319
Query 80 ANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGDVLLNR 139
A AFGV EFI+ E+S FIA+GR CK+D+VN I+ +RPD ++ Y I++GD+LLNR
Sbjct 320 AEAFGVGVEFIDQELSRFIAAGRLHCKIDKVNEIVETNRPDSKNWQYQETIKKGDLLLNR 379
Query 140 IQKLARVL 147
+QKL+RV+
Sbjct 380 VQKLSRVI 387
> 7300859
Length=389
Score = 132 bits (333), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 60/133 (45%), Positives = 92/133 (69%), Gaps = 0/133 (0%)
Query 15 DMKGLLNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSV 74
D+K L + Y +Y F +L + K+++ D + PHY Y++R +R+ Y Q L+ Y+S+
Sbjct 255 DVKQFLFSLYNCQYENFYVHLAGVEKQLRLDYLIHPHYRYYVREMRILGYTQLLESYRSL 314
Query 75 TLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGD 134
TL MA +FGV+ E+I+ E++ FIA+GR KVDRV GI+ +RPD+++ Y I+QGD
Sbjct 315 TLQYMAESFGVTVEYIDQELARFIAAGRLHAKVDRVGGIVETNRPDNKNWQYQATIKQGD 374
Query 135 VLLNRIQKLARVL 147
+LLNRIQKL+RV+
Sbjct 375 LLLNRIQKLSRVI 387
> At4g24820
Length=406
Score = 122 bits (305), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 55/110 (50%), Positives = 81/110 (73%), Gaps = 0/110 (0%)
Query 38 IAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLINMANAFGVSPEFIEAEVSSF 97
+A ++K D YL PH+ +++R +R Y Q+L+ YKSVT+ MA AFGVS +FI+ E+S F
Sbjct 295 MAVQIKYDRYLYPHFRFYMREVRTVVYSQFLESYKSVTVEAMAKAFGVSVDFIDQELSRF 354
Query 98 IASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGDVLLNRIQKLARVL 147
IA+G+ CK+D+V G++ +RPD ++ Y I+QGD LLNRIQKL+RV+
Sbjct 355 IAAGKLHCKIDKVAGVLETNRPDAKNALYQATIKQGDFLLNRIQKLSRVI 404
> CE03368
Length=410
Score = 120 bits (301), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 60/129 (46%), Positives = 86/129 (66%), Gaps = 1/129 (0%)
Query 20 LNAFYFAKY-REFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLIN 78
L ++Y Y R F+Q ++R K D YLSPH+ Y+ R +R RAY+Q+L PYK+V +
Sbjct 280 LESYYDCHYDRFFIQLAALESERFKFDRYLSPHFNYYSRGMRHRAYEQFLTPYKTVRIDM 339
Query 79 MANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGDVLLN 138
MA FGVS FI+ E+ IA+G+ C++D VNG+I + D ++ Y +I+ GD+LLN
Sbjct 340 MAKDFGVSRAFIDRELHRLIATGQLQCRIDAVNGVIEVNHRDSKNHLYKAVIKDGDILLN 399
Query 139 RIQKLARVL 147
RIQKLARV+
Sbjct 400 RIQKLARVI 408
> SPBC582.07c
Length=409
Score = 114 bits (284), Expect = 8e-26, Method: Compositional matrix adjust.
Identities = 48/128 (37%), Positives = 86/128 (67%), Gaps = 1/128 (0%)
Query 16 MKGLLNAFYFAKYREFMQYLVPI-AKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSV 74
++ +N+ Y Y F + L + +K D +L HY Y++R +R RAY Q L+ Y+++
Sbjct 270 LEACINSLYLCDYSGFFRTLADVEVNHLKCDQFLVAHYRYYVREMRRRAYAQLLESYRAL 329
Query 75 TLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGD 134
++ +MA +FGVS ++I+ +++SFI + C +DRVNG++ +RPD+++ Y +++QGD
Sbjct 330 SIDSMAASFGVSVDYIDRDLASFIPDNKLNCVIDRVNGVVFTNRPDEKNRQYQEVVKQGD 389
Query 135 VLLNRIQK 142
VLLN++QK
Sbjct 390 VLLNKLQK 397
> YPR108w
Length=429
Score = 95.1 bits (235), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 47/133 (35%), Positives = 81/133 (60%), Gaps = 1/133 (0%)
Query 11 GAGEDMKGLLNAFYFAKYREFMQYLVPIAKRVKTDI-YLSPHYLYFIRSIRLRAYQQYLD 69
A + + L + Y + Y + YL+ V YL+ H +F+R +R + Y Q L+
Sbjct 284 AALQSISSLTISLYASDYASYFPYLLETYANVLIPCKYLNRHADFFVREMRRKVYAQLLE 343
Query 70 PYKSVTLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPI 129
YK+++L +MA+AFGVS F++ ++ FI + + C +DRVNGI+ +RPD+++ Y +
Sbjct 344 SYKTLSLKSMASAFGVSVAFLDNDLGKFIPNKQLNCVIDRVNGIVETNRPDNKNAQYHLL 403
Query 130 IRQGDVLLNRIQK 142
++QGD LL ++QK
Sbjct 404 VKQGDGLLTKLQK 416
> ECU10g0190
Length=381
Score = 75.1 bits (183), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 38/127 (29%), Positives = 69/127 (54%), Gaps = 2/127 (1%)
Query 19 LLNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLIN 78
L + + Y +FM L K ++ D+++ F R +RLR Y Q L+ Y+ + L N
Sbjct 257 LATSLFECNYGDFMNDLYLFCKSLQDDVFVGRFVDLFCREMRLRVYGQVLESYRLMLLEN 316
Query 79 MANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGDVLLN 138
MA F VS E++E ++ FI GR K+DR++G++ +++ +S ++ G ++
Sbjct 317 MAQTFRVSVEYVERDLGEFIVEGRLWSKIDRISGVVEVTSREEQDVS--SVVGYGCDVVR 374
Query 139 RIQKLAR 145
RI+K +
Sbjct 375 RIKKCVK 381
> At3g61140
Length=440
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 36/120 (30%), Positives = 63/120 (52%), Gaps = 0/120 (0%)
Query 15 DMKGLLNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSV 74
D++ L+N FY ++Y ++YL + + DI+L H IR +A QY P+ SV
Sbjct 293 DVRELINDFYSSRYASCLEYLASLKSNLLLDIHLHDHVDTLYDQIRKKALIQYTLPFVSV 352
Query 75 TLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGD 134
L MA+AF S +E E+ + I + ++D N I+ A D R+ ++ +++ G+
Sbjct 353 DLSRMADAFKTSVSGLEKELEALITDNQIQARIDSHNKILYARHADQRNATFQKVLQMGN 412
> 7293847
Length=525
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 35/111 (31%), Positives = 54/111 (48%), Gaps = 0/111 (0%)
Query 16 MKGLLNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVT 75
++ ++ FY +KY + L I + D+Y++PH IR RA QY PY S
Sbjct 355 LRDIIFKFYESKYASCLTLLDEIRDNLLVDMYIAPHVTTLYTKIRNRALIQYFSPYMSAD 414
Query 76 LINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSY 126
+ MA AF S +E EV I G+ ++D N I+ A D R+ ++
Sbjct 415 MHKMAMAFNSSVGDLENEVMQLILDGQIQARIDSHNKILFAKEADQRNSTF 465
> Hs5713317
Length=500
Score = 58.5 bits (140), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 34/104 (32%), Positives = 51/104 (49%), Gaps = 0/104 (0%)
Query 23 FYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLINMANA 82
FY +KY ++ L + + D+YL+PH IR RA QY PY S + MA A
Sbjct 342 FYESKYASCLKMLDEMKDNLLLDMYLAPHVRTLYTQIRNRALIQYFSPYVSADMHRMAAA 401
Query 83 FGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSY 126
F + +E E++ I G +VD + I+ A D RS ++
Sbjct 402 FNTTVAALEDELTQLILEGLISARVDSHSKILYARDVDQRSTTF 445
> 7297479
Length=444
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 25/111 (22%), Positives = 52/111 (46%), Gaps = 0/111 (0%)
Query 16 MKGLLNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVT 75
M L+N++ EF L + D ++ H +R+IR + + + PYK++
Sbjct 312 MTNLVNSYQNNDINEFETILRQHRSNIMADQFIREHIEDLLRNIRTQVLIKLIRPYKNIA 371
Query 76 LINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSY 126
+ +ANA + P +E+ + S I ++D+VN ++ + + + Y
Sbjct 372 IPFIANALNIEPAEVESLLVSCILDDTIKGRIDQVNQVLQLDKINSSASRY 422
> CE26200
Length=601
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 28/129 (21%), Positives = 59/129 (45%), Gaps = 4/129 (3%)
Query 19 LLNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLIN 78
LL ++ +++ + + + R+ D ++S + IR + QYL PY ++ +
Sbjct 398 LLGSYTSSRFGRCFEIMRSVKPRLLLDPFISRNVDELFEKIRQKCVLQYLQPYSTIKMAT 457
Query 79 MANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGDVLLN 138
MA A G+S ++ + I K+D+ GI+ D + I+++ +V +
Sbjct 458 MAEAVGMSSAELQLSLLELIEQKHVSLKIDQNEGIVRILDERDEN----AILKRVNVTCD 513
Query 139 RIQKLARVL 147
R + A+ L
Sbjct 514 RATQKAKSL 522
> SPBC215.03c
Length=422
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 37/84 (44%), Gaps = 0/84 (0%)
Query 27 KYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLINMANAFGVS 86
KY + L A+ D+YL+P IR R+ YL PY ++ +A F +
Sbjct 275 KYSLLLDTLQQNAQDYSLDMYLAPQLTNLFSLIRERSLLDYLIPYSALPFSKIAVDFHID 334
Query 87 PEFIEAEVSSFIASGRPVCKVDRV 110
FIE + I + + KVD +
Sbjct 335 ENFIEKNLLEIIEAKKLNGKVDSL 358
> Hs4506221
Length=456
Score = 34.3 bits (77), Expect = 0.091, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 71 YKSVTLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDD 121
Y +T+ MA +S + EA +S+ + + KVDR+ GII RP D
Sbjct 370 YTRITMKRMAQLLDLSVDESEAFLSNLVVNKTIFAKVDRLAGIINFQRPKD 420
> Hs7019469
Length=1359
Score = 32.0 bits (71), Expect = 0.39, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 71 YKSVTLINMANAFGVSPEFIEAEVSSFIASGRP 103
Y +V+LIN+ +A G+S EF+ SF S +P
Sbjct 1190 YNAVSLINLVSAVGMSVEFVSHITRSFAISTKP 1222
> 7298174
Length=364
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 19/36 (52%), Gaps = 0/36 (0%)
Query 16 MKGLLNAFYFAKYREFMQYLVPIAKRVKTDIYLSPH 51
M +L FY ++ M L I V+ D+YLSPH
Sbjct 298 MWTILAKFYAGEFDACMTLLREIENHVRLDVYLSPH 333
> 7297621
Length=1157
Score = 30.4 bits (67), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 24/38 (63%), Gaps = 1/38 (2%)
Query 72 KSVTLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDR 109
+V+L+N+ A G+S EF V SF A+ + V ++DR
Sbjct 1029 NAVSLVNLVMAVGISVEFCSHLVHSF-ATSKSVSQIDR 1065
> At3g13930
Length=508
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 19/88 (21%), Positives = 42/88 (47%), Gaps = 24/88 (27%)
Query 28 YREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLINMANAFGVSP 87
++EF++Y++ + R++ +L Y YF Y YL P++ + + ++ SP
Sbjct 53 FKEFIRYVIHLPMRIQLATFL---YEYF--------YPNYLPPHQEIGMPSL------SP 95
Query 88 EFIEAEVSSF-------IASGRPVCKVD 108
E ++ + +A G +C+V+
Sbjct 96 TMTEGNIARWLKKEGDKVAPGEVLCEVE 123
> 7294586
Length=575
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 19/54 (35%), Positives = 28/54 (51%), Gaps = 7/54 (12%)
Query 85 VSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGDVLLN 138
+ P+F+++ S FIA C D V G I RP DR + P+ G VL++
Sbjct 137 ILPKFVQS--SGFIA-----CVHDLVKGSIMLRRPIDRRHVHAPLPNVGAVLMH 183
> Hs18554904
Length=113
Score = 28.5 bits (62), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 25/51 (49%), Gaps = 0/51 (0%)
Query 71 YKSVTLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDD 121
Y +T+ M +S + EA +S+ + + KVDR+ GI RP D
Sbjct 59 YTRMTMKRMVQLLDLSVDESEAFLSNLVVNKTIFAKVDRLAGINNFQRPKD 109
> At5g09900
Length=442
Score = 28.5 bits (62), Expect = 4.7, Method: Composition-based stats.
Identities = 14/44 (31%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 71 YKSVTLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGII 114
Y +TL +A +S E E +S + S + K+DR +GI+
Sbjct 353 YARITLKRLAELLCLSMEEAEKHLSEMVVSKALIAKIDRPSGIV 396
> Hs18573626_1
Length=653
Score = 28.5 bits (62), Expect = 4.8, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 42 VKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLIN 78
+K DI + LY I++ LR ++Y+ KS+T IN
Sbjct 61 LKRDIAMLKQELYAIKNDSLRKEKEYIHEIKSITEIN 97
> Hs4759264
Length=443
Score = 27.7 bits (60), Expect = 8.3, Method: Composition-based stats.
Identities = 18/99 (18%), Positives = 43/99 (43%), Gaps = 0/99 (0%)
Query 16 MKGLLNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVT 75
M L++A+ EF + L + D ++ H +R+IR + + + PY +
Sbjct 311 MTNLVSAYQNNDITEFEKILKTNHSNIMDDPFIREHIEELLRNIRTQVLIKLIKPYTRIH 370
Query 76 LINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGII 114
+ ++ + +E+ + I ++D+VN ++
Sbjct 371 IPFISKELNIDVADVESLLVQCILDNTIHGRIDQVNQLL 409
> At5g64760
Length=529
Score = 27.3 bits (59), Expect = 9.4, Method: Composition-based stats.
Identities = 13/44 (29%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 71 YKSVTLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGII 114
Y +T +A ++ E E +S + S + K+DR +GII
Sbjct 440 YSRITFKRLAELLCLTTEEAEKHLSEMVVSKALIAKIDRPSGII 483
Lambda K H
0.326 0.141 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1785281974
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40