bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1375_orf1
Length=67
Score E
Sequences producing significant alignments: (Bits) Value
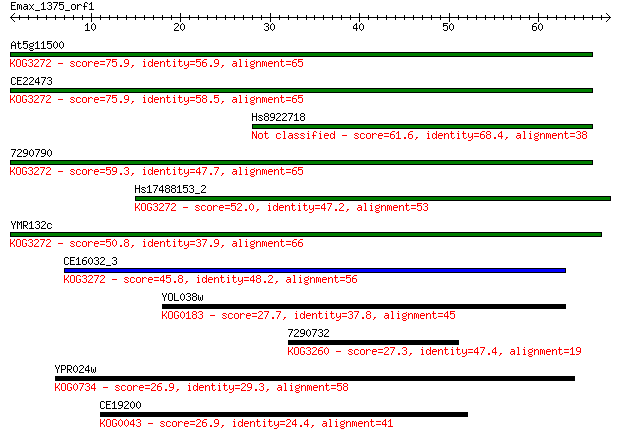
At5g11500 75.9 2e-14
CE22473 75.9 2e-14
Hs8922718 61.6 3e-10
7290790 59.3 2e-09
Hs17488153_2 52.0 2e-07
YMR132c 50.8 6e-07
CE16032_3 45.8 2e-05
YOL038w 27.7 5.1
7290732 27.3 6.5
YPR024w 26.9 9.0
CE19200 26.9 9.7
> At5g11500
Length=215
Score = 75.9 bits (185), Expect = 2e-14, Method: Composition-based stats.
Identities = 37/65 (56%), Positives = 48/65 (73%), Gaps = 1/65 (1%)
Query 1 AHVYVRMPYGQTDYESLPPAVIEEACQLTKHNSIEGCKQSEVTIVYTPASNLKKTASMDT 60
AHVY+R+ GQ+ ++ + V+E+ QL K NSI+G K + V +VYTP SNLKKTASMD
Sbjct 47 AHVYLRLHRGQS-FDDISEGVLEDCAQLVKANSIQGNKVNNVDVVYTPWSNLKKTASMDV 105
Query 61 GQVGF 65
GQVGF
Sbjct 106 GQVGF 110
> CE22473
Length=210
Score = 75.9 bits (185), Expect = 2e-14, Method: Composition-based stats.
Identities = 38/65 (58%), Positives = 46/65 (70%), Gaps = 1/65 (1%)
Query 1 AHVYVRMPYGQTDYESLPPAVIEEACQLTKHNSIEGCKQSEVTIVYTPASNLKKTASMDT 60
AHVY+R+ G T +S+P A++ + CQL K NSIEGCK + V IVYT SNLKKT M
Sbjct 47 AHVYLRLHSGMT-IDSIPEALLIDCCQLVKQNSIEGCKLNNVAIVYTMWSNLKKTGDMAV 105
Query 61 GQVGF 65
GQVGF
Sbjct 106 GQVGF 110
> Hs8922718
Length=140
Score = 61.6 bits (148), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 26/38 (68%), Positives = 30/38 (78%), Gaps = 0/38 (0%)
Query 28 LTKHNSIEGCKQSEVTIVYTPASNLKKTASMDTGQVGF 65
L K NSI+GCK + V +VYTP SNLKKTA MD GQ+GF
Sbjct 6 LVKANSIQGCKMNNVNVVYTPWSNLKKTADMDVGQIGF 43
> 7290790
Length=209
Score = 59.3 bits (142), Expect = 2e-09, Method: Composition-based stats.
Identities = 31/65 (47%), Positives = 43/65 (66%), Gaps = 1/65 (1%)
Query 1 AHVYVRMPYGQTDYESLPPAVIEEACQLTKHNSIEGCKQSEVTIVYTPASNLKKTASMDT 60
AHVY+R+ GQT + +P V+ +A QL K NSI+G K + + +VYT NLKKT M+
Sbjct 47 AHVYLRLRKGQT-IDDIPTDVLVDAAQLCKANSIQGNKVNNLEVVYTMWENLKKTPDMEP 105
Query 61 GQVGF 65
GQV +
Sbjct 106 GQVAY 110
> Hs17488153_2
Length=238
Score = 52.0 bits (123), Expect = 2e-07, Method: Composition-based stats.
Identities = 25/53 (47%), Positives = 34/53 (64%), Gaps = 1/53 (1%)
Query 15 ESLPPAVIEEACQLTKHNSIEGCKQSEVTIVYTPASNLKKTASMDTGQVGFKQ 67
E +P V+ + L K NS +GCK + V +V TP SNLKKTA +D Q+GF +
Sbjct 5 EDIPKEVLTDCAHLVKANSTQGCKNN-VNVVNTPRSNLKKTADVDVRQIGFHR 56
> YMR132c
Length=208
Score = 50.8 bits (120), Expect = 6e-07, Method: Composition-based stats.
Identities = 25/66 (37%), Positives = 38/66 (57%), Gaps = 0/66 (0%)
Query 1 AHVYVRMPYGQTDYESLPPAVIEEACQLTKHNSIEGCKQSEVTIVYTPASNLKKTASMDT 60
HVY+++ + + +P VI + QL K SI+G K + TI+ TP NL+K M+
Sbjct 51 GHVYLKLRPNEKTIDDIPQEVICDCLQLCKSESIQGNKMPQCTILITPWHNLRKNRYMNP 110
Query 61 GQVGFK 66
G+V FK
Sbjct 111 GEVSFK 116
> CE16032_3
Length=148
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 27/71 (38%), Positives = 33/71 (46%), Gaps = 15/71 (21%)
Query 7 MPYGQTDYESLPPAVI---------------EEACQLTKHNSIEGCKQSEVTIVYTPASN 51
M +G PPA+I EE CQL K NSI+G K +V + YT N
Sbjct 1 MVFGFISSTVTPPALIFMGEHQVENEKLFKLEECCQLVKKNSIQGVKMEKVEVNYTLKEN 60
Query 52 LKKTASMDTGQ 62
LKK M TG+
Sbjct 61 LKKVKGMQTGE 71
> YOL038w
Length=254
Score = 27.7 bits (60), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 27/49 (55%), Gaps = 5/49 (10%)
Query 18 PPAVIEEACQLTKHNSIE----GCKQSEVTIVYTPASNLKKTASMDTGQ 62
PPA +EE +LT + +E G K E+T+V P S++ +S + Q
Sbjct 184 PPATVEECVKLTVRSLLEVVQTGAKNIEITVV-KPDSDIVALSSEEINQ 231
> 7290732
Length=230
Score = 27.3 bits (59), Expect = 6.5, Method: Composition-based stats.
Identities = 9/19 (47%), Positives = 13/19 (68%), Gaps = 0/19 (0%)
Query 32 NSIEGCKQSEVTIVYTPAS 50
N ++GC + VT+ YTP S
Sbjct 87 NGVQGCTEENVTVTYTPNS 105
> YPR024w
Length=747
Score = 26.9 bits (58), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 28/58 (48%), Gaps = 3/58 (5%)
Query 6 RMPYGQTDYESLPPAVIEEACQLTKHNSIEGCKQSEVTIVYTPASNLKKTASMDTGQV 63
R+ G +YE+L IE+ C K ++ K S T+V P S+ +K D ++
Sbjct 687 RLAQGLIEYETLDAHEIEQVC---KGEKLDKLKTSTNTVVEGPDSDERKDIGDDKPKI 741
> CE19200
Length=478
Score = 26.9 bits (58), Expect = 9.7, Method: Composition-based stats.
Identities = 10/41 (24%), Positives = 19/41 (46%), Gaps = 0/41 (0%)
Query 11 QTDYESLPPAVIEEACQLTKHNSIEGCKQSEVTIVYTPASN 51
Q ++ + PP ++ C L N GC+ + +Y S+
Sbjct 243 QREFINRPPMLVYRCCWLDHMNDFHGCRMKRIFKMYVYCSD 283
Lambda K H
0.312 0.127 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160559522
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40