bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1366_orf1
Length=122
Score E
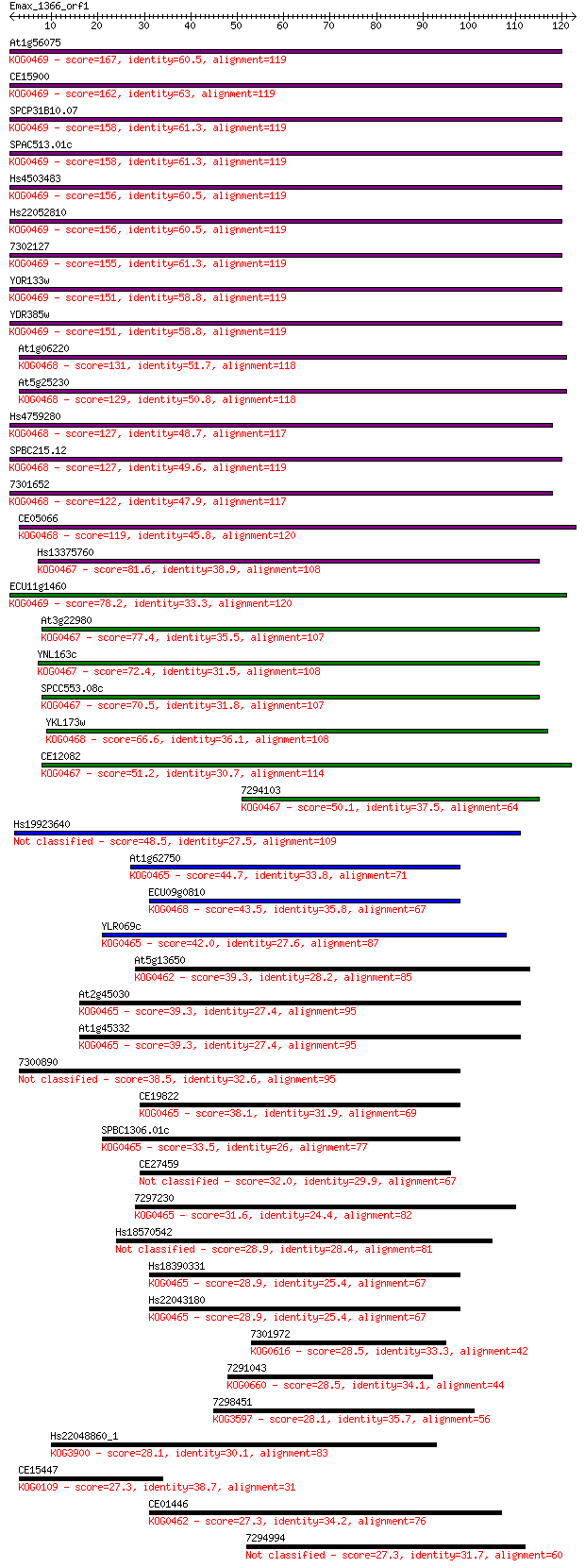
Sequences producing significant alignments: (Bits) Value
At1g56075 167 5e-42
CE15900 162 2e-40
SPCP31B10.07 158 2e-39
SPAC513.01c 158 2e-39
Hs4503483 156 8e-39
Hs22052810 156 8e-39
7302127 155 1e-38
YOR133w 151 2e-37
YDR385w 151 2e-37
At1g06220 131 2e-31
At5g25230 129 1e-30
Hs4759280 127 5e-30
SPBC215.12 127 7e-30
7301652 122 1e-28
CE05066 119 2e-27
Hs13375760 81.6 3e-16
ECU11g1460 78.2 4e-15
At3g22980 77.4 7e-15
YNL163c 72.4 2e-13
SPCC553.08c 70.5 8e-13
YKL173w 66.6 1e-11
CE12082 51.2 5e-07
7294103 50.1 1e-06
Hs19923640 48.5 3e-06
At1g62750 44.7 4e-05
ECU09g0810 43.5 9e-05
YLR069c 42.0 3e-04
At5g13650 39.3 0.002
At2g45030 39.3 0.002
At1g45332 39.3 0.002
7300890 38.5 0.003
CE19822 38.1 0.004
SPBC1306.01c 33.5 0.099
CE27459 32.0 0.32
7297230 31.6 0.43
Hs18570542 28.9 2.1
Hs18390331 28.9 2.2
Hs22043180 28.9 2.6
7301972 28.5 2.8
7291043 28.5 3.0
7298451 28.1 4.5
Hs22048860_1 28.1 4.5
CE15447 27.3 6.3
CE01446 27.3 7.5
7294994 27.3 7.9
> At1g56075
Length=855
Score = 167 bits (422), Expect = 5e-42, Method: Compositional matrix adjust.
Identities = 72/119 (60%), Positives = 96/119 (80%), Gaps = 0/119 (0%)
Query 1 DAIHRGAGQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVF 60
DAIHRG GQ++PT RRV+YA+Q+ ++PRL EP+++V+I P ++ GI +VL +RGHVF
Sbjct 709 DAIHRGGGQVIPTARRVIYASQITAKPRLLEPVYMVEIQAPEGALGGIYSVLNQKRGHVF 768
Query 61 EEDSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPLEKGSK 119
EE +PG PL ++AYLPV ESFGF++ LRAATSGQAFPQCVFDHW ++ DPLE G++
Sbjct 769 EEMQRPGTPLYNIKAYLPVVESFGFSSQLRAATSGQAFPQCVFDHWEMMSSDPLEPGTQ 827
> CE15900
Length=852
Score = 162 bits (409), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 75/119 (63%), Positives = 89/119 (74%), Gaps = 0/119 (0%)
Query 1 DAIHRGAGQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVF 60
DAIHRG GQI+PT RRV YA+ L ++PRL EP++LV+I P ++ GI VL RRGHVF
Sbjct 706 DAIHRGGGQIIPTARRVFYASVLTAEPRLLEPVYLVEIQCPEAAVGGIYGVLNRRRGHVF 765
Query 61 EEDSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPLEKGSK 119
EE G P+ V++AYLPV ESFGFT LR+ T GQAFPQCVFDHW L GDPLE G+K
Sbjct 766 EESQVTGTPMFVVKAYLPVNESFGFTADLRSNTGGQAFPQCVFDHWQVLPGDPLEAGTK 824
> SPCP31B10.07
Length=842
Score = 158 bits (400), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 73/119 (61%), Positives = 93/119 (78%), Gaps = 0/119 (0%)
Query 1 DAIHRGAGQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVF 60
DAIHRG GQI+PT RRV+YA+ L++ P +QEP+FLV+I V ++M GI +VL +RGHVF
Sbjct 696 DAIHRGGGQIIPTARRVVYASTLLASPIIQEPVFLVEIQVSENAMGGIYSVLNKKRGHVF 755
Query 61 EEDSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPLEKGSK 119
E+ + G PL ++AYLPV ESFGFT LR AT+GQAFPQ VFDHWS ++GDPL+ SK
Sbjct 756 SEEQRVGTPLYNIKAYLPVNESFGFTGELRQATAGQAFPQLVFDHWSPMSGDPLDPTSK 814
> SPAC513.01c
Length=812
Score = 158 bits (400), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 73/119 (61%), Positives = 93/119 (78%), Gaps = 0/119 (0%)
Query 1 DAIHRGAGQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVF 60
DAIHRG GQI+PT RRV+YA+ L++ P +QEP+FLV+I V ++M GI +VL +RGHVF
Sbjct 666 DAIHRGGGQIIPTARRVVYASTLLASPIIQEPVFLVEIQVSENAMGGIYSVLNKKRGHVF 725
Query 61 EEDSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPLEKGSK 119
E+ + G PL ++AYLPV ESFGFT LR AT+GQAFPQ VFDHWS ++GDPL+ SK
Sbjct 726 SEEQRVGTPLYNIKAYLPVNESFGFTGELRQATAGQAFPQLVFDHWSPMSGDPLDPTSK 784
> Hs4503483
Length=858
Score = 156 bits (395), Expect = 8e-39, Method: Compositional matrix adjust.
Identities = 72/119 (60%), Positives = 86/119 (72%), Gaps = 0/119 (0%)
Query 1 DAIHRGAGQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVF 60
DAIHRG GQI+PT RR LYA+ L +QPRL EP++LV+I P + GI VL +RGHVF
Sbjct 712 DAIHRGGGQIIPTARRCLYASVLTAQPRLMEPIYLVEIQCPEQVVGGIYGVLNRKRGHVF 771
Query 61 EEDSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPLEKGSK 119
EE G P+ V++AYLPV ESFGFT LR+ T GQAFPQCVFDHW L GDP + S+
Sbjct 772 EESQVAGTPMFVVKAYLPVNESFGFTADLRSNTGGQAFPQCVFDHWQILPGDPFDNSSR 830
> Hs22052810
Length=846
Score = 156 bits (395), Expect = 8e-39, Method: Compositional matrix adjust.
Identities = 72/119 (60%), Positives = 86/119 (72%), Gaps = 0/119 (0%)
Query 1 DAIHRGAGQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVF 60
DAIHRG GQI+PT RR LYA+ L +QPRL EP++LV+I P + GI VL +RGHVF
Sbjct 700 DAIHRGGGQIIPTARRCLYASVLTAQPRLMEPIYLVEIQCPEQVVGGIYGVLNRKRGHVF 759
Query 61 EEDSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPLEKGSK 119
EE G P+ V++AYLPV ESFGFT LR+ T GQAFPQCVFDHW L GDP + S+
Sbjct 760 EESQVAGTPMFVVKAYLPVNESFGFTADLRSNTGGQAFPQCVFDHWQILPGDPFDNSSR 818
> 7302127
Length=832
Score = 155 bits (393), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 73/119 (61%), Positives = 87/119 (73%), Gaps = 0/119 (0%)
Query 1 DAIHRGAGQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVF 60
DAIHRG GQI+PT RR LYAA + ++PRL EP++L +I P ++ GI VL RRGHVF
Sbjct 686 DAIHRGGGQIIPTTRRCLYAAAITAKPRLMEPVYLCEIQCPEVAVGGIYGVLNRRRGHVF 745
Query 61 EEDSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPLEKGSK 119
EE+ G P+ V++AYLPV ESFGFT LR+ T GQAFPQCVFDHW L GDP E SK
Sbjct 746 EENQVVGTPMFVVKAYLPVNESFGFTADLRSNTGGQAFPQCVFDHWQVLPGDPSEPSSK 804
> YOR133w
Length=842
Score = 151 bits (382), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 70/119 (58%), Positives = 86/119 (72%), Gaps = 0/119 (0%)
Query 1 DAIHRGAGQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVF 60
DAIHRG GQI+PT RR YA L++ P++QEP+FLV+I P ++ GI +VL +RG V
Sbjct 696 DAIHRGGGQIIPTMRRATYAGFLLADPKIQEPVFLVEIQCPEQAVGGIYSVLNKKRGQVV 755
Query 61 EEDSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPLEKGSK 119
E+ +PG PL ++AYLPV ESFGFT LR AT GQAFPQ VFDHWS L DPL+ SK
Sbjct 756 SEEQRPGTPLFTVKAYLPVNESFGFTGELRQATGGQAFPQMVFDHWSTLGSDPLDPTSK 814
> YDR385w
Length=842
Score = 151 bits (382), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 70/119 (58%), Positives = 86/119 (72%), Gaps = 0/119 (0%)
Query 1 DAIHRGAGQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVF 60
DAIHRG GQI+PT RR YA L++ P++QEP+FLV+I P ++ GI +VL +RG V
Sbjct 696 DAIHRGGGQIIPTMRRATYAGFLLADPKIQEPVFLVEIQCPEQAVGGIYSVLNKKRGQVV 755
Query 61 EEDSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPLEKGSK 119
E+ +PG PL ++AYLPV ESFGFT LR AT GQAFPQ VFDHWS L DPL+ SK
Sbjct 756 SEEQRPGTPLFTVKAYLPVNESFGFTGELRQATGGQAFPQMVFDHWSTLGSDPLDPTSK 814
> At1g06220
Length=987
Score = 131 bits (330), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 61/118 (51%), Positives = 83/118 (70%), Gaps = 0/118 (0%)
Query 3 IHRGAGQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVFEE 62
+HRG+GQ++PT RRV Y+A L++ PRL EP++ V+I P D + I TVL RRGHV +
Sbjct 815 LHRGSGQMIPTARRVAYSAFLMATPRLMEPVYYVEIQTPIDCVTAIYTVLSRRRGHVTSD 874
Query 63 DSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPLEKGSKM 120
+PG P +++A+LPV ESFGF T LR T GQAF VFDHW+ + GDPL+K ++
Sbjct 875 VPQPGTPAYIVKAFLPVIESFGFETDLRYHTQGQAFCLSVFDHWAIVPGDPLDKAIQL 932
> At5g25230
Length=973
Score = 129 bits (325), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 60/118 (50%), Positives = 83/118 (70%), Gaps = 0/118 (0%)
Query 3 IHRGAGQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVFEE 62
+HRG+GQ++PT RRV Y+A L++ PRL EP++ V+I P D + I TVL RRG+V +
Sbjct 801 LHRGSGQMIPTARRVAYSAFLMATPRLMEPVYYVEIQTPIDCVTAIYTVLSRRRGYVTSD 860
Query 63 DSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPLEKGSKM 120
+PG P +++A+LPV ESFGF T LR T GQAF VFDHW+ + GDPL+K ++
Sbjct 861 VPQPGTPAYIVKAFLPVIESFGFETDLRYHTQGQAFCLSVFDHWAIVPGDPLDKAIQL 918
> Hs4759280
Length=972
Score = 127 bits (319), Expect = 5e-30, Method: Compositional matrix adjust.
Identities = 57/117 (48%), Positives = 79/117 (67%), Gaps = 0/117 (0%)
Query 1 DAIHRGAGQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVF 60
+ +HRG GQI+PT RRV+Y+A L++ PRL EP + V++ P D + + TVL RRGHV
Sbjct 799 EPLHRGGGQIIPTARRVVYSAFLMATPRLMEPYYFVEVQAPADCVSAVYTVLARRRGHVT 858
Query 61 EEDSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPLEKG 117
++ PG+PL ++A++P +SFGF T LR T GQAF VF HW + GDPL+K
Sbjct 859 QDAPIPGSPLYTIKAFIPAIDSFGFETDLRTHTQGQAFSLSVFHHWQIVPGDPLDKS 915
> SPBC215.12
Length=983
Score = 127 bits (318), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 59/119 (49%), Positives = 82/119 (68%), Gaps = 0/119 (0%)
Query 1 DAIHRGAGQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVF 60
+ I+RG GQI+PT RRV Y++ L + PRL EP+++V++ P DS+ I +L RRGHV
Sbjct 812 EQIYRGGGQIIPTARRVCYSSFLTASPRLMEPVYMVEVHAPADSLPIIYDLLTRRRGHVL 871
Query 61 EEDSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPLEKGSK 119
++ +PG+PL ++RA +PV +S GF T LR T GQA Q VFDHW + GDPL+K K
Sbjct 872 QDIPRPGSPLYLVRALIPVIDSCGFETDLRVHTQGQAMCQMVFDHWQVVPGDPLDKSIK 930
> 7301652
Length=975
Score = 122 bits (307), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 56/117 (47%), Positives = 77/117 (65%), Gaps = 0/117 (0%)
Query 1 DAIHRGAGQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVF 60
+A+HRG GQI+PT RRV Y+A L++ PRL EP V++ P D + + TVL RRGHV
Sbjct 802 EALHRGGGQIIPTARRVAYSAFLMATPRLMEPYLFVEVQAPADCVSAVYTVLARRRGHVT 861
Query 61 EEDSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPLEKG 117
++ G+P+ ++A++P +SFGF T LR T GQAF VF HW + GDPL+K
Sbjct 862 QDAPVSGSPIYTIKAFIPAIDSFGFETDLRTHTQGQAFCLSVFHHWQIVPGDPLDKS 918
> CE05066
Length=974
Score = 119 bits (297), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 55/120 (45%), Positives = 77/120 (64%), Gaps = 0/120 (0%)
Query 3 IHRGAGQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVFEE 62
++RG GQ++PT RR Y+A L++ PRL EP + V++ P D + + TVL RRGHV +
Sbjct 805 LYRGGGQMIPTARRCAYSAFLMATPRLMEPYYTVEVVAPADCVAAVYTVLAKRRGHVTTD 864
Query 63 DSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPLEKGSKMET 122
PG+P+ + AY+PV +SFGF T LR T GQAF F HW + GDPL+K ++T
Sbjct 865 APMPGSPMYTISAYIPVMDSFGFETDLRIHTQGQAFCMSAFHHWQLVPGDPLDKSIVIKT 924
> Hs13375760
Length=857
Score = 81.6 bits (200), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 42/108 (38%), Positives = 57/108 (52%), Gaps = 0/108 (0%)
Query 7 AGQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVFEEDSKP 66
+GQ++ T + A V RL M+ DI D + + VL R G V +E+ K
Sbjct 695 SGQLIATMKEACRYALQVKPQRLMAAMYTCDIMATGDVLGRVYAVLSKREGRVLQEEMKE 754
Query 67 GNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPL 114
G + +++A LPVAESFGF +R TSG A PQ VF HW + DP
Sbjct 755 GTDMFIIKAVLPVAESFGFADEIRKRTSGLASPQLVFSHWEIIPSDPF 802
> ECU11g1460
Length=850
Score = 78.2 bits (191), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 40/120 (33%), Positives = 63/120 (52%), Gaps = 0/120 (0%)
Query 1 DAIHRGAGQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVF 60
DAIHRG Q++ + + L + P L EP++ V+I+ P D + T+L +RG
Sbjct 704 DAIHRGINQLLQPVKNLCKGLLLAAGPILYEPIYEVEITTPNDYSGAVTTILLSKRGTAE 763
Query 61 EEDSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPLEKGSKM 120
+ + PGN ++ LPV ESF F L++ + G+A F H+S L G+ + S M
Sbjct 764 DFKTLPGNDTTMITGTLPVKESFTFNEDLKSGSRGKAGASMRFSHYSILPGNLEDPNSLM 823
> At3g22980
Length=1015
Score = 77.4 bits (189), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 38/107 (35%), Positives = 57/107 (53%), Gaps = 0/107 (0%)
Query 8 GQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVFEEDSKPG 67
GQ+M + AA L + PR+ E M+ +++ + + + VL RR + +E+ + G
Sbjct 853 GQVMTAVKDACRAAVLQTNPRIVEAMYFCELNTAPEYLGPMYAVLSRRRARILKEEMQEG 912
Query 68 NPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPL 114
+ L + AY+PV+ESFGF LR TSG A V HW L DP
Sbjct 913 SSLFTVHAYVPVSESFGFADELRKGTSGGASALMVLSHWEMLEEDPF 959
> YNL163c
Length=1110
Score = 72.4 bits (176), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 34/108 (31%), Positives = 58/108 (53%), Gaps = 0/108 (0%)
Query 7 AGQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVFEEDSKP 66
+G+++ + R ++ A L PR+ ++ DI D + + V+ R G + E+ K
Sbjct 949 SGRLITSTRDAIHEAFLDWSPRIMWAIYSCDIQTSVDVLGKVYAVILQRHGKIISEEMKE 1008
Query 67 GNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPL 114
G P + A++PV E+FG + +R TSG A PQ VF + C++ DP
Sbjct 1009 GTPFFQIEAHVPVVEAFGLSEDIRKRTSGAAQPQLVFSGFECIDLDPF 1056
> SPCC553.08c
Length=1000
Score = 70.5 bits (171), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 34/107 (31%), Positives = 56/107 (52%), Gaps = 0/107 (0%)
Query 8 GQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVFEEDSKPG 67
GQ++ + + L PRL M+ D+ + + + V+ RRG V +E+ K G
Sbjct 841 GQVISVVKESIRHGFLGWSPRLMLAMYSCDVQATSEVLGRVYGVVSKRRGRVIDEEMKEG 900
Query 68 NPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPL 114
P +++A +PV ESFGF + TSG A+PQ +F + L+ +P
Sbjct 901 TPFFIVKALIPVVESFGFAVEILKRTSGAAYPQLIFHGFEMLDENPF 947
> YKL173w
Length=1008
Score = 66.6 bits (161), Expect = 1e-11, Method: Composition-based stats.
Identities = 39/111 (35%), Positives = 61/111 (54%), Gaps = 3/111 (2%)
Query 9 QIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRG-HVFEEDSKPG 67
QI+P ++ Y L + P L EP++ VDI+V + ++ ++ RRG +++ G
Sbjct 837 QIIPLMKKACYVGLLTAIPILLEPIYEVDITVHAPLLPIVEELMKKRRGSRIYKTIKVAG 896
Query 68 NPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDH--WSCLNGDPLEK 116
PL+ +R +PV ES GF T LR +T+G Q F H W + GD L+K
Sbjct 897 TPLLEVRGQVPVIESAGFETDLRLSTNGLGMCQLYFWHKIWRKVPGDVLDK 947
> CE12082
Length=906
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 35/126 (27%), Positives = 50/126 (39%), Gaps = 12/126 (9%)
Query 8 GQIMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHV-------- 59
GQ+M + AA RL M+ ++ ++ + VL R+ V
Sbjct 733 GQMMTAIKASCSAAAKKLALRLVAAMYRCTVTTASQALGKVHAVLSQRKSKVGMENCRTF 792
Query 60 ----FEEDSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLNGDPLE 115
ED L + + +PV ESF F LR TSG A Q F HW ++ DP
Sbjct 793 SNIVLSEDINEATNLFEVVSLMPVVESFSFCDQLRKFTSGMASAQLQFSHWQVIDEDPYW 852
Query 116 KGSKME 121
S +E
Sbjct 853 TPSTLE 858
> 7294103
Length=122
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 32/64 (50%), Gaps = 0/64 (0%)
Query 51 VLFVRRGHVFEEDSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLN 110
V+ R G + D G+ + LPV ESF F +R TSG A PQ +F HW ++
Sbjct 4 VIGRRHGKILSGDLTQGSGNFAVTCLLPVIESFNFAQEMRKQTSGLACPQLMFSHWEVID 63
Query 111 GDPL 114
DP
Sbjct 64 IDPF 67
> Hs19923640
Length=779
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 30/111 (27%), Positives = 55/111 (49%), Gaps = 4/111 (3%)
Query 2 AIHRGAGQIMPTC--RRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHV 59
IH G M + R + A + ++ EP+ ++++V RD + + L RRG++
Sbjct 656 TIHPGTSTTMISACVSRCVQKALKKADKQVLEPLMNLEVTVARDYLSPVLADLAQRRGNI 715
Query 60 FEEDSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSCLN 110
E ++ N +V+ ++P+AE G++T LR TSG A + +N
Sbjct 716 QEIQTRQDNKVVI--GFVPLAEIMGYSTVLRTLTSGSATFALELSTYQAMN 764
> At1g62750
Length=783
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 39/71 (54%), Gaps = 1/71 (1%)
Query 27 PRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVFEEDSKPGNPLVVLRAYLPVAESFGFT 86
PR+ EP+ V++ P + + + L RRG + KPG L V+ + +P+AE F +
Sbjct 685 PRMLEPIMRVEVVTPEEHLGDVIGDLNSRRGQINSFGDKPGG-LKVVDSLVPLAEMFQYV 743
Query 87 TALRAATSGQA 97
+ LR T G+A
Sbjct 744 STLRGMTKGRA 754
> ECU09g0810
Length=615
Score = 43.5 bits (101), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 36/67 (53%), Gaps = 0/67 (0%)
Query 31 EPMFLVDISVPRDSMEGIDTVLFVRRGHVFEEDSKPGNPLVVLRAYLPVAESFGFTTALR 90
EP++LV+I+ +D+ + + V+ G V + P + L YLPV ESFGF T LR
Sbjct 515 EPLYLVEITHAKDAEDLVSEVISSSFGEVIHQSRFPFSTLESTLCYLPVPESFGFETDLR 574
Query 91 AATSGQA 97
+ A
Sbjct 575 VYSCSTA 581
> YLR069c
Length=761
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/87 (27%), Positives = 47/87 (54%), Gaps = 2/87 (2%)
Query 21 AQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVFEEDSKPGNPLVVLRAYLPVA 80
A L +QP + EP+ V ++ P + +G + + + + +D++ G+ L+A ++
Sbjct 656 AFLRAQPVIMEPIMNVSVTSP-NEFQG-NVIGLLNKLQAVIQDTENGHDEFTLKAECALS 713
Query 81 ESFGFTTALRAATSGQAFPQCVFDHWS 107
FGF T+LRA+T G+ F H++
Sbjct 714 TMFGFATSLRASTQGKGEFSLEFSHYA 740
> At5g13650
Length=609
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 35/85 (41%), Gaps = 0/85 (0%)
Query 28 RLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVFEEDSKPGNPLVVLRAYLPVAESFGFTT 87
+L EP + + VP M + +L RRG +F+ LR +P G
Sbjct 406 KLLEPYEIATVEVPEAHMGPVVELLGKRRGQMFDMQGVGSEGTTFLRYKIPTRGLLGLRN 465
Query 88 ALRAATSGQAFPQCVFDHWSCLNGD 112
A+ A+ G A VFD + GD
Sbjct 466 AILTASRGTAILNTVFDSYGPWAGD 490
> At2g45030
Length=754
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 50/96 (52%), Gaps = 6/96 (6%)
Query 16 RVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVFEEDSKPGNPLVVLRA 75
R+ Y A ++P + EP+ LV++ VP + + + R+G + D + + V+ A
Sbjct 647 RLCYTA---ARPVILEPVMLVELKVPTEFQGTVAGDINKRKGIIVGNDQEGDDS--VITA 701
Query 76 YLPVAESFGFTTALRAATSGQA-FPQCVFDHWSCLN 110
+P+ FG++T+LR+ T G+ F +H + N
Sbjct 702 NVPLNNMFGYSTSLRSMTQGKGEFTMEYKEHSAVSN 737
> At1g45332
Length=754
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 50/96 (52%), Gaps = 6/96 (6%)
Query 16 RVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVFEEDSKPGNPLVVLRA 75
R+ Y A ++P + EP+ LV++ VP + + + R+G + D + + V+ A
Sbjct 647 RLCYTA---ARPVILEPVMLVELKVPTEFQGTVAGDINKRKGIIVGNDQEGDDS--VITA 701
Query 76 YLPVAESFGFTTALRAATSGQA-FPQCVFDHWSCLN 110
+P+ FG++T+LR+ T G+ F +H + N
Sbjct 702 NVPLNNMFGYSTSLRSMTQGKGEFTMEYKEHSAVSN 737
> 7300890
Length=1093
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 31/100 (31%), Positives = 48/100 (48%), Gaps = 9/100 (9%)
Query 3 IHRGAGQ--IMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVF 60
I RG +M T + + S RL EP+ + I P + + GI L RR +
Sbjct 569 IGRGTADSFVMATAAQCVQKLLSTSGTRLLEPIMALQIVAPSERISGIMADLSRRRALI- 627
Query 61 EEDSKPG---NPLVVLRAYLPVAESFGFTTALRAATSGQA 97
D P N ++++ A P+AE G+++ALR +SG A
Sbjct 628 -NDVLPKGERNKMILVNA--PLAELSGYSSALRTISSGTA 664
> CE19822
Length=750
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 36/69 (52%), Gaps = 2/69 (2%)
Query 29 LQEPMFLVDISVPRDSMEGIDTVLFVRRGHVFEEDSKPGNPLVVLRAYLPVAESFGFTTA 88
L EP+ V+ +VP + + T L R + DS G V+ A P+++ FG+T+
Sbjct 642 LLEPIMKVEATVPTEFQGNVVTSLTQRNALITTTDSTEGYATVICEA--PLSDMFGYTSE 699
Query 89 LRAATSGQA 97
LR+ T G+
Sbjct 700 LRSLTEGKG 708
> SPBC1306.01c
Length=558
Score = 33.5 bits (75), Expect = 0.099, Method: Compositional matrix adjust.
Identities = 20/77 (25%), Positives = 38/77 (49%), Gaps = 2/77 (2%)
Query 21 AQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVFEEDSKPGNPLVVLRAYLPVA 80
A L + P + EP+ V I+ P + G+ L R+ + + D+ L+A +P+
Sbjct 454 AFLQANPMVLEPIMNVSITAPVEHQGGVIGNLDKRKATIVDSDTDEDE--FTLQAEVPLN 511
Query 81 ESFGFTTALRAATSGQA 97
F +++ +RA T G+
Sbjct 512 SMFSYSSDIRALTKGKG 528
> CE27459
Length=689
Score = 32.0 bits (71), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 31/68 (45%), Gaps = 1/68 (1%)
Query 29 LQEPMFLVDISVPRDS-MEGIDTVLFVRRGHVFEEDSKPGNPLVVLRAYLPVAESFGFTT 87
L EP+ V I + D + I L RR H D+ + + A LP++E+ +
Sbjct 592 LTEPVMEVQIEIRNDDPTQPILNELLRRRAHFEHSDATESTEIRRICAILPLSETENLSK 651
Query 88 ALRAATSG 95
+R TSG
Sbjct 652 TVRTLTSG 659
> 7297230
Length=729
Score = 31.6 bits (70), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 20/83 (24%), Positives = 40/83 (48%), Gaps = 3/83 (3%)
Query 28 RLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVFEEDSKPGNPLVVLRAYLPVAESFGFTT 87
++ EP+ LV+++ P + + L R G + + G + A +P+ + FG+
Sbjct 622 QILEPIMLVEVTAPEEFQGAVMGHLSKRHGIITGTEGTEG--WFTVYAEVPLNDMFGYAG 679
Query 88 ALRAATSGQAFPQCVFDHWS-CL 109
LR++T G+ + +S CL
Sbjct 680 ELRSSTQGKGEFTMEYSRYSPCL 702
> Hs18570542
Length=421
Score = 28.9 bits (63), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 39/93 (41%), Gaps = 17/93 (18%)
Query 24 VSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVFEEDSKPGNPLVVLRAYLPV---- 79
+QP +V ++ P I ++ G + D+ P NPL+V+ + V
Sbjct 8 ATQPSEANSFVIVALAFPL-----ISALITTMSGFIPRGDTMPQNPLLVVNGEIAVTGYT 62
Query 80 -AESFGFTTALRA-------ATSGQAFPQCVFD 104
A S F T L A+SG+++P+ F
Sbjct 63 WATSINFITPLMTAGMCNAPASSGKSYPESKFS 95
> Hs18390331
Length=751
Score = 28.9 bits (63), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 17/67 (25%), Positives = 32/67 (47%), Gaps = 2/67 (2%)
Query 31 EPMFLVDISVPRDSMEGIDTVLFVRRGHVFEEDSKPGNPLVVLRAYLPVAESFGFTTALR 90
EP+ V++ P + + + R G + +D L A +P+ + FG++T LR
Sbjct 647 EPIMAVEVVAPNEFQGQVIAGINRRHGVITGQDGV--EDYFTLYADVPLNDMFGYSTELR 704
Query 91 AATSGQA 97
+ T G+
Sbjct 705 SCTEGKG 711
> Hs22043180
Length=485
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 17/67 (25%), Positives = 32/67 (47%), Gaps = 2/67 (2%)
Query 31 EPMFLVDISVPRDSMEGIDTVLFVRRGHVFEEDSKPGNPLVVLRAYLPVAESFGFTTALR 90
EP+ V++ P + + + R G + +D L A +P+ + FG++T LR
Sbjct 381 EPIMAVEVVAPNEFQGQVIAGINRRHGVITGQDGV--EDYFTLYADVPLNDMFGYSTELR 438
Query 91 AATSGQA 97
+ T G+
Sbjct 439 SCTEGKG 445
> 7301972
Length=393
Score = 28.5 bits (62), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Query 53 FVRRGHVFEEDSKPGNPLVVLRAYLPVAESFGFTTALRAATS 94
++ + H+ D KP N L+ R Y+ + + FGFT + TS
Sbjct 158 YMHKMHLMYRDLKPENILLDQRGYIKITD-FGFTKRVDGRTS 198
> 7291043
Length=392
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 23/44 (52%), Gaps = 1/44 (2%)
Query 48 IDTVLFVRRGHVFEEDSKPGNPLVVLRAYLPVAESFGFTTALRA 91
I+ + F+ G+V D KP N L+ + L VA+ FG L +
Sbjct 134 INAIKFIHSGNVIHRDLKPSNILIDSKCRLKVAD-FGLARTLSS 176
> 7298451
Length=2225
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 29/56 (51%), Gaps = 2/56 (3%)
Query 45 MEGIDTVLFVRRGHVFEEDSKPGNPLVVLRAYLPVAESFGFTTALRAATSGQAFPQ 100
+EGI+ VL V ++ + DS+PGN + + E+FG A TS AF Q
Sbjct 451 IEGIERVLDVDLFNIDDPDSEPGNLIYTILPTQSPQETFGCFIVGGATTS--AFSQ 504
> Hs22048860_1
Length=1121
Score = 28.1 bits (61), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 25/85 (29%), Positives = 35/85 (41%), Gaps = 4/85 (4%)
Query 10 IMPTCRRVLYAAQLVSQPRLQEPMFLVDISVPRDSMEGIDTVLFVRRGHVFEEDSKPGNP 69
+ P RV Y + + +P L P +S+ R + D V R + K G
Sbjct 373 VAPLGSRVTYRSTVQGRPPLANPPETKGVSLKR--LGAPDRTGPVVRSCPHKRTPKWGKK 430
Query 70 LVV--LRAYLPVAESFGFTTALRAA 92
++ LRA LP ESFG L A
Sbjct 431 RILRPLRAALPARESFGLAQGLSLA 455
> CE15447
Length=305
Score = 27.3 bits (59), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 12/32 (37%), Positives = 20/32 (62%), Gaps = 1/32 (3%)
Query 3 IHRGAGQ-IMPTCRRVLYAAQLVSQPRLQEPM 33
+ R AG I+ + +R+ Y AQ V +P + +PM
Sbjct 124 VKRSAGDPIIDSAKRIAYGAQSVVEPEIPQPM 155
> CE01446
Length=645
Score = 27.3 bits (59), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 35/77 (45%), Gaps = 2/77 (2%)
Query 31 EPMFLVDISVPRDSMEGIDTVLFVRRGHVFEEDSKPGNPLVVLRAYLPVAE-SFGFTTAL 89
EPM V + VP + M ++ + RG E S LV+L LP+AE + F L
Sbjct 445 EPMVKVRMIVPNEMMGTVNGLCSECRGERGEITSIDSTRLVIL-WRLPLAEVAVDFFERL 503
Query 90 RAATSGQAFPQCVFDHW 106
+ TSG A D W
Sbjct 504 KKLTSGYASFDYEPDGW 520
> 7294994
Length=516
Score = 27.3 bits (59), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 27/63 (42%), Gaps = 3/63 (4%)
Query 52 LFVRRGHVFEEDSKPG---NPLVVLRAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSC 108
L G F D PG +P ++ + +A+ T A+R TSG +P FD C
Sbjct 156 LNCANGQAFIMDCAPGTAFSPASLVCVHKDLAKCGSGTGAVRDDTSGTGYPALPFDDLGC 215
Query 109 LNG 111
G
Sbjct 216 PPG 218
Lambda K H
0.323 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194805952
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40