bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
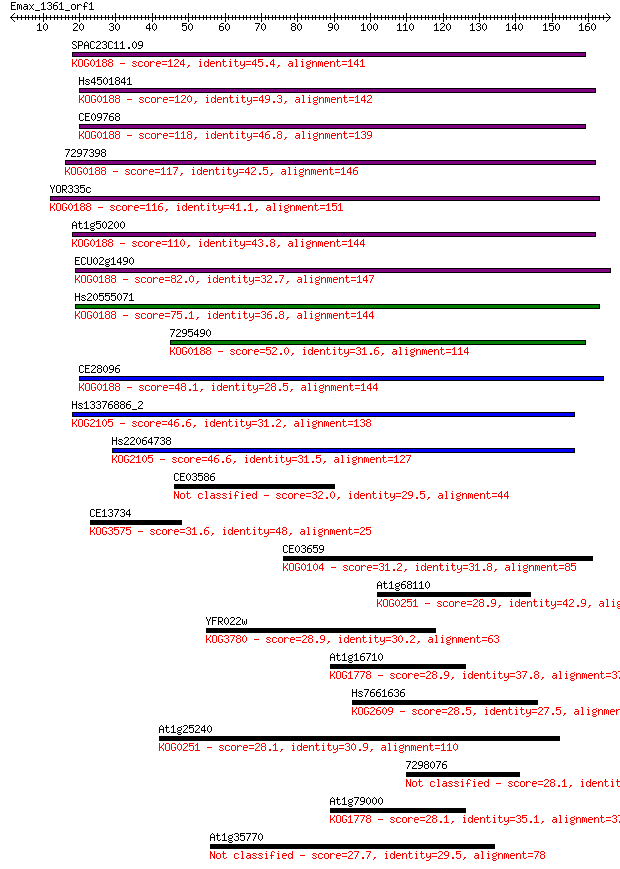
Query= Emax_1361_orf1
Length=165
Score E
Sequences producing significant alignments: (Bits) Value
SPAC23C11.09 124 1e-28
Hs4501841 120 1e-27
CE09768 118 6e-27
7297398 117 9e-27
YOR335c 116 2e-26
At1g50200 110 9e-25
ECU02g1490 82.0 4e-16
Hs20555071 75.1 7e-14
7295490 52.0 5e-07
CE28096 48.1 8e-06
Hs13376886_2 46.6 2e-05
Hs22064738 46.6 2e-05
CE03586 32.0 0.52
CE13734 31.6 0.80
CE03659 31.2 0.86
At1g68110 28.9 4.2
YFR022w 28.9 5.0
At1g16710 28.9 5.4
Hs7661636 28.5 5.8
At1g25240 28.1 7.3
7298076 28.1 7.3
At1g79000 28.1 7.9
At1g35770 27.7 9.9
> SPAC23C11.09
Length=959
Score = 124 bits (310), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 64/141 (45%), Positives = 86/141 (60%), Gaps = 2/141 (1%)
Query 18 IKQKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPA 77
I Q+GSLV + +LRFDF + + + ++ +E Y I +L V + + L A E+
Sbjct 624 IDQRGSLVSQEKLRFDFSYKSSIPIDKLLLVENYCNNVIQDNLSVYSKEVALSKAKEING 683
Query 78 LCAVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVV 137
L AVFGEVYPDPVRVV IG D L +QE + D S+E CGGTH + +++DFV+
Sbjct 684 LRAVFGEVYPDPVRVVCIGVDIDTL--LQEPKKPDWTKYSIEFCGGTHCDKSGEIKDFVI 741
Query 138 ISEESIAKGIRRIVAVAGEAA 158
+ E IAKGIRRIVAV A
Sbjct 742 LEENGIAKGIRRIVAVTSTEA 762
> Hs4501841
Length=968
Score = 120 bits (301), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 70/142 (49%), Positives = 83/142 (58%), Gaps = 2/142 (1%)
Query 20 QKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPALC 79
QKGSLV RLRFDF ++ QQI+ E I ++ V T PL A + L
Sbjct 624 QKGSLVAPDRLRFDFTAKGAMSTQQIKKAEEIANEMIEAAKAVYTQDCPLAAAKAIQGLR 683
Query 80 AVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVVIS 139
AVF E YPDPVRVVSIG EL + + G SVE CGGTHL N+S FV+++
Sbjct 684 AVFDETYPDPVRVVSIGVPVSEL--LDDPSGPAGSLTSVEFCGGTHLRNSSHAGAFVIVT 741
Query 140 EESIAKGIRRIVAVAGEAAVSA 161
EE+IAKGIRRIVAV G A A
Sbjct 742 EEAIAKGIRRIVAVTGAEAQKA 763
> CE09768
Length=968
Score = 118 bits (295), Expect = 6e-27, Method: Compositional matrix adjust.
Identities = 65/139 (46%), Positives = 85/139 (61%), Gaps = 2/139 (1%)
Query 20 QKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPALC 79
QKGSLV R+RFDF A +T QQ++ E Y Q+ I + V PL A ++ L
Sbjct 625 QKGSLVAPDRMRFDFTNKAGMTVQQVKKAEEYAQQLIDTKGQVYAKNSPLGEAKKVKGLR 684
Query 80 AVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVVIS 139
A+F E YPDPVRVV++G ++L +Q + N +VE CGGTHL N S + V+ S
Sbjct 685 AMFDETYPDPVRVVAVGTPVEQL--LQNPDAEEGQNTTVEFCGGTHLQNVSHIGRIVIAS 742
Query 140 EESIAKGIRRIVAVAGEAA 158
EE+IAKGIRRIVA+ G A
Sbjct 743 EEAIAKGIRRIVALTGPEA 761
> 7297398
Length=966
Score = 117 bits (294), Expect = 9e-27, Method: Compositional matrix adjust.
Identities = 62/146 (42%), Positives = 90/146 (61%), Gaps = 2/146 (1%)
Query 16 RTIKQKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMEL 75
+ +QKGSLV +LRFDF+ A +T +Q+ E + + ++P+ L A ++
Sbjct 620 KDTEQKGSLVVPEKLRFDFNSKAAMTIEQVSKTEQLTKEMVYKNVPIYAKESKLALAKKI 679
Query 76 PALCAVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDF 135
L +VF EVYPDPVRV+S G EL+ +++ ++ SVE CGGTHL + + DF
Sbjct 680 RGLRSVFDEVYPDPVRVISFGVPVDELEQNPDSEAGEQ--TSVEFCGGTHLRRSGHIMDF 737
Query 136 VVISEESIAKGIRRIVAVAGEAAVSA 161
V+ SEE+IAKGIRRIVA+ G A+ A
Sbjct 738 VISSEEAIAKGIRRIVALTGPEALKA 763
> YOR335c
Length=958
Score = 116 bits (290), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 62/151 (41%), Positives = 87/151 (57%), Gaps = 2/151 (1%)
Query 12 ETERRTIKQKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKT 71
ET + QKGSLV +LRFDF ++N++++ +E I +L V +PL
Sbjct 612 ETLGNDVDQKGSLVAPEKLRFDFSHKKAVSNEELKKVEDICNEQIKENLQVFYKEIPLDL 671
Query 72 AMELPALCAVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQ 131
A + + AVFGE YPDPVRVVS+G +EL + + S+E CGGTH++
Sbjct 672 AKSIDGVRAVFGETYPDPVRVVSVGKPIEEL--LANPANEEWTKYSIEFCGGTHVNKTGD 729
Query 132 LEDFVVISEESIAKGIRRIVAVAGEAAVSAR 162
++ FV++ E IAKGIRRIVAV G A A+
Sbjct 730 IKYFVILEESGIAKGIRRIVAVTGTEAFEAQ 760
> At1g50200
Length=1027
Score = 110 bits (276), Expect = 9e-25, Method: Compositional matrix adjust.
Identities = 63/144 (43%), Positives = 84/144 (58%), Gaps = 2/144 (1%)
Query 18 IKQKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPA 77
I QKGS+V +LRFDF P+ + + IE V + I L V + L A +
Sbjct 688 IDQKGSIVLPEKLRFDFSHGKPVDPEDLRRIESIVNKQIKDELDVFSKEAVLSEAKRIKG 747
Query 78 LCAVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVV 137
L AVFGEVYPDPVRVVSIG ++L + + + + S E CGGTH++N + + F +
Sbjct 748 LRAVFGEVYPDPVRVVSIGRKVEDL--LADPENNEWSLLSSEFCGGTHITNTREAKAFAL 805
Query 138 ISEESIAKGIRRIVAVAGEAAVSA 161
+SEE IAKGIRR+ AV E A A
Sbjct 806 LSEEGIAKGIRRVTAVTTECAFDA 829
> ECU02g1490
Length=864
Score = 82.0 bits (201), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 48/149 (32%), Positives = 81/149 (54%), Gaps = 18/149 (12%)
Query 19 KQKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPAL 78
+Q+GSLV+ +LRFDF+ LTN+++E +E + I V +L + AM+ P +
Sbjct 570 QQRGSLVDPDKLRFDFE-GRKLTNEELEDVERKINDFIKKGAEVKAVILDRRQAMDNPNI 628
Query 79 CAVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVVI 138
+ E YP+ VR++++ D ++ E+CGGTH++N S + F ++
Sbjct 629 VRMQNEEYPENVRIITMQSDGLIIQ---------------EMCGGTHVTNVSDIGKFRIV 673
Query 139 SEESIAKGIRRIVAVAGEAA--VSARSEL 165
SE ++ RRIV V G+ A + A +EL
Sbjct 674 SETGVSANTRRIVGVTGKKAHELDAEAEL 702
> Hs20555071
Length=985
Score = 75.1 bits (183), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 53/144 (36%), Positives = 77/144 (53%), Gaps = 4/144 (2%)
Query 19 KQKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPAL 78
+Q+GS + +LR D PLT +Q+ A+E VQ A+ V +PL ++P L
Sbjct 651 EQQGSHLNPEQLRLDVTTQTPLTPEQLRAVENTVQEAVGQDEAVYMEEVPLALTAQVPGL 710
Query 79 CAVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVVI 138
++ EVYPDPVRVVS+ + + + SVELC GTHL + D V+I
Sbjct 711 RSL-DEVYPDPVRVVSV---GVPVAHALDPASQAALQTSVELCCGTHLLRTGAVGDLVII 766
Query 139 SEESIAKGIRRIVAVAGEAAVSAR 162
+ ++KG R++AV GE A AR
Sbjct 767 GDRQLSKGTTRLLAVTGEQAQQAR 790
> 7295490
Length=1012
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 36/115 (31%), Positives = 60/115 (52%), Gaps = 14/115 (12%)
Query 45 IEAIELYVQRAIASSLPVNTAVLPLKTAMELPALCAVFGEVYPDP-VRVVSIGPDTKELK 103
++ IE + R I S+ PV +L +E + V GEVYP+ +R+V++ ++ EL+
Sbjct 702 VQLIEDLINRVICSAAPVEVQLLSAAEVLEQNDITMVPGEVYPEQGLRLVNV--ESPELQ 759
Query 104 TIQETQGADKINASVELCGGTHLSNASQLEDFVVISEESIAKGIRRIVAVAGEAA 158
+S ELC GTH +N S+L F +++ + + AVAG+AA
Sbjct 760 L-----------SSKELCCGTHATNTSELSCFCIVNLKQTNRARFAFTAVAGQAA 803
> CE28096
Length=793
Score = 48.1 bits (113), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 41/151 (27%), Positives = 73/151 (48%), Gaps = 22/151 (14%)
Query 20 QKGSLVEERRLRFDFD-WDAPLTNQQ----IEAIELYVQRAIASSLPVNTAVLPLKTAME 74
QKGS V+ R RFD+ D L+ +Q + E+ ++ I + L+ A +
Sbjct 613 QKGSSVDCDRFRFDYSTGDEDLSKEQRTELLIKCEMKMREFIQNGGFTEIIETSLEEAKK 672
Query 75 LPALCAVFGE--VYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQL 132
+ L + E + VRVV++G GAD VE C GTH+ + +
Sbjct 673 IENLQSDVKEDRIGGASVRVVALGS------------GAD---VPVECCSGTHIHDVRVI 717
Query 133 EDFVVISEESIAKGIRRIVAVAGEAAVSARS 163
+D ++S++S+ + +RRI+ + G+ A + R+
Sbjct 718 DDVAIMSDKSMGQRLRRIIVLTGKEAAACRN 748
> Hs13376886_2
Length=428
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 43/143 (30%), Positives = 61/143 (42%), Gaps = 27/143 (18%)
Query 18 IKQKGSLVEERRLRFDFDWDAP-LTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELP 76
K K + E R R + D P +T +Q+ AIE V I LPVN L L P
Sbjct 139 FKLKTTSWELGRFRSAIELDTPSMTAEQVAAIEQSVNEKIRDRLPVNVRELSLDD----P 194
Query 77 ALCAVFGEVYPD----PVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQL 132
+ V G PD P+RVV+I +G D +C GTH+SN S L
Sbjct 195 EVEQVSGRGLPDDHAGPIRVVNI-------------EGVDS-----NMCCGTHVSNLSDL 236
Query 133 EDFVVISEESIAKGIRRIVAVAG 155
+ ++ E K ++ ++G
Sbjct 237 QVIKILGTEKGKKNRTNLIFLSG 259
> Hs22064738
Length=307
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 40/132 (30%), Positives = 57/132 (43%), Gaps = 27/132 (20%)
Query 29 RLRFDFDWDAP-LTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPALCAVFGEVYP 87
R R + D P +T +Q+ AIE V I LPVN L L P + V G P
Sbjct 29 RFRSAIELDTPSMTAEQVAAIEQSVNEKIRDRLPVNVRELSLDD----PEVEQVSGRGLP 84
Query 88 D----PVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVVISEESI 143
D P+RVV+I +G D +C GTH+SN S L+ ++ E
Sbjct 85 DDHAGPIRVVNI-------------EGVDS-----NMCCGTHVSNLSDLQVIKILGTEKG 126
Query 144 AKGIRRIVAVAG 155
K ++ ++G
Sbjct 127 KKNRTNLIFLSG 138
> CE03586
Length=207
Score = 32.0 bits (71), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 13/44 (29%), Positives = 24/44 (54%), Gaps = 0/44 (0%)
Query 46 EAIELYVQRAIASSLPVNTAVLPLKTAMELPALCAVFGEVYPDP 89
++YV +IAS+ + T + ++ M +P LCA+ + DP
Sbjct 57 SFFQIYVMDSIASTFLLLTDMFFVRLFMYVPPLCAIISPFFWDP 100
> CE13734
Length=511
Score = 31.6 bits (70), Expect = 0.80, Method: Composition-based stats.
Identities = 12/25 (48%), Positives = 19/25 (76%), Gaps = 0/25 (0%)
Query 23 SLVEERRLRFDFDWDAPLTNQQIEA 47
SLV + ++FDFDW + L+N Q++A
Sbjct 362 SLVNTQTVKFDFDWLSHLSNAQMKA 386
> CE03659
Length=926
Score = 31.2 bits (69), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 42/86 (48%), Gaps = 9/86 (10%)
Query 76 PALCAVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINAS-VELCGGTHLSNASQLED 134
P ++FGE YP P RV+ + + K + Q ADK S VE+ G + +A + E
Sbjct 457 PIRKSLFGENYPVPNRVMHFSSYSDDFKI--DIQDADKNPLSTVEISG---VKDAIEKE- 510
Query 135 FVVISEESIAKGIRRIVAVAGEAAVS 160
V E S+ KG++ ++ VS
Sbjct 511 --VTDENSVLKGVKTTFSIDLSGIVS 534
> At1g68110
Length=379
Score = 28.9 bits (63), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 24/44 (54%), Gaps = 3/44 (6%)
Query 102 LKTIQET--QGADKINASVELCGGTHLSNASQLEDFVVISEESI 143
LK + +T QG D I E C G +SNA ++ FV I EE +
Sbjct 243 LKIVNKTTSQGEDLI-VYFEFCKGFGVSNAREIPQFVRIPEEEV 285
> YFR022w
Length=733
Score = 28.9 bits (63), Expect = 5.0, Method: Composition-based stats.
Identities = 19/65 (29%), Positives = 31/65 (47%), Gaps = 7/65 (10%)
Query 55 AIASSLPVNTAVLPLKTAMELPALCAVFGEVYP--DPVRVVSIGPDTKELKTIQETQGAD 112
AI S+ P+N +++PL ++L ++ V E Y DP P E + + E D
Sbjct 248 AIGSATPINISIVPLSKGLKLGSIKVVLFENYQYCDP-----FPPVISENRQVTELNLED 302
Query 113 KINAS 117
+N S
Sbjct 303 PLNES 307
> At1g16710
Length=1706
Score = 28.9 bits (63), Expect = 5.4, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 89 PVRVVSIGPDTKELKTIQETQGADKINASVELCGGTH 125
PV + I DTK+ I E++ D A + LC G H
Sbjct 1465 PVEIADIPTDTKDRDEILESEFFDTRQAFLSLCQGNH 1501
> Hs7661636
Length=243
Score = 28.5 bits (62), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 14/51 (27%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 95 IGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVVISEESIAK 145
I PD + + ++E G + S L GTH+ + +++ V+ E+ I K
Sbjct 142 IKPDMETYERLREKHGEEFFPTSNSLLHGTHVPSTEEIDRMVIDLEKQIEK 192
> At1g25240
Length=376
Score = 28.1 bits (61), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 34/126 (26%), Positives = 56/126 (44%), Gaps = 19/126 (15%)
Query 42 NQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPALCAVFGEVYPDPVRVVS------- 94
NQ++E IE +Q + L + +K + L A+ V E++ R+ S
Sbjct 166 NQELERIE-KLQSLLHMLLQIRPMADNMKKTLILEAMDCVVIEIFDIYGRICSAIAKLLI 224
Query 95 -IGPDTKE------LKTIQE--TQGADKINASVELCGGTHLSNASQLEDFVVISEESIAK 145
I P + LK +++ +QG D + E C +SNA + FV I EE I K
Sbjct 225 KIHPAAGKAEAVIALKIVKKATSQGED-LALYFEFCKEFGVSNAHDIPKFVTIPEEDI-K 282
Query 146 GIRRIV 151
I +++
Sbjct 283 AIEKVI 288
> 7298076
Length=301
Score = 28.1 bits (61), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 16/31 (51%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 110 GADKINASVELCGGTHLSNASQLEDFVVISE 140
GA + AS +L HLSNA +LE+ VISE
Sbjct 45 GATETEASAQLDPQEHLSNADRLENNRVISE 75
> At1g79000
Length=1691
Score = 28.1 bits (61), Expect = 7.9, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 89 PVRVVSIGPDTKELKTIQETQGADKINASVELCGGTH 125
PV ++ I DT++ I E++ D A + LC G H
Sbjct 1450 PVEIMDIPADTRDKDEILESEFFDTRQAFLSLCQGNH 1486
> At1g35770
Length=968
Score = 27.7 bits (60), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 36/78 (46%), Gaps = 8/78 (10%)
Query 56 IASSLPVNTAVLPLKTAMELPALCAVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKIN 115
I +P +A LP+ T +PA C P R V++ P+ + Q + K
Sbjct 711 IVVPIPDVSADLPVTT---VPAFCRGAPSAVSSP-RQVNVDPEER----FQNSMKKLKAI 762
Query 116 ASVELCGGTHLSNASQLE 133
+S+ +CGG LSN L+
Sbjct 763 SSISICGGIALSNKDILD 780
Lambda K H
0.313 0.130 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2353551590
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40