bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1310_orf2
Length=151
Score E
Sequences producing significant alignments: (Bits) Value
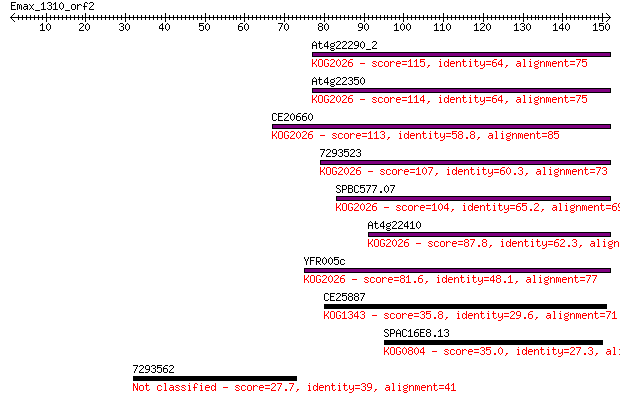
At4g22290_2 115 3e-26
At4g22350 114 6e-26
CE20660 113 1e-25
7293523 107 7e-24
SPBC577.07 104 8e-23
At4g22410 87.8 6e-18
YFR005c 81.6 5e-16
CE25887 35.8 0.034
SPAC16E8.13 35.0 0.050
7293562 27.7 8.9
> At4g22290_2
Length=543
Score = 115 bits (288), Expect = 3e-26, Method: Composition-based stats.
Identities = 48/75 (64%), Positives = 60/75 (80%), Gaps = 0/75 (0%)
Query 77 VRRTCPYLGTINKHMLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMHALEQR 136
VRR CPYL T+N+ +LDFDFE+ CS+ LSN +VYACLVCG+YFQGR + + A+ H+LE
Sbjct 107 VRRDCPYLDTVNRQVLDFDFERFCSVSLSNLNVYACLVCGKYFQGRSQKSHAYTHSLEAG 166
Query 137 HFVYVNLTTCKVYCL 151
H VY+NL T KVYCL
Sbjct 167 HHVYINLLTEKVYCL 181
> At4g22350
Length=477
Score = 114 bits (286), Expect = 6e-26, Method: Composition-based stats.
Identities = 48/75 (64%), Positives = 60/75 (80%), Gaps = 0/75 (0%)
Query 77 VRRTCPYLGTINKHMLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMHALEQR 136
VRR CPYL T+N+ +LDFDFE+ CS+ LSN +VYACLVCG+YFQGR + + A+ H+LE
Sbjct 105 VRRDCPYLDTVNRQVLDFDFERFCSVSLSNLNVYACLVCGKYFQGRSQKSHAYTHSLEAG 164
Query 137 HFVYVNLTTCKVYCL 151
H VY+NL T KVYCL
Sbjct 165 HHVYINLLTEKVYCL 179
> CE20660
Length=602
Score = 113 bits (283), Expect = 1e-25, Method: Composition-based stats.
Identities = 50/85 (58%), Positives = 64/85 (75%), Gaps = 1/85 (1%)
Query 67 SSSSAPVQKRVRRTCPYLGTINKHMLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRST 126
S A +K+ R CPYL TI++ +LDFDFEK CS+ LS+Q+VYAC+VCG+YFQGRG +T
Sbjct 123 SMKKAQAEKK-SRMCPYLDTIDRSVLDFDFEKQCSVSLSHQNVYACMVCGKYFQGRGTNT 181
Query 127 FAFMHALEQRHFVYVNLTTCKVYCL 151
A+ HALE H V++NL T K YCL
Sbjct 182 HAYTHALETDHHVFLNLQTLKFYCL 206
> 7293523
Length=719
Score = 107 bits (268), Expect = 7e-24, Method: Composition-based stats.
Identities = 44/73 (60%), Positives = 59/73 (80%), Gaps = 0/73 (0%)
Query 79 RTCPYLGTINKHMLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMHALEQRHF 138
R CPYL TIN+++LDFDFEK+CSI L+ +VYACLVCG+YFQGRG +T A+ H++ + H
Sbjct 25 RVCPYLDTINRNLLDFDFEKLCSISLTRINVYACLVCGKYFQGRGTNTHAYTHSVGEAHH 84
Query 139 VYVNLTTCKVYCL 151
V++NL T + YCL
Sbjct 85 VFLNLHTLRFYCL 97
> SPBC577.07
Length=502
Score = 104 bits (259), Expect = 8e-23, Method: Composition-based stats.
Identities = 45/69 (65%), Positives = 54/69 (78%), Gaps = 0/69 (0%)
Query 83 YLGTINKHMLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMHALEQRHFVYVN 142
YL TIN+ +LDFDFEKVCS+ L+N VYACLVCGRYFQGRG S+ A+ HAL + H V+VN
Sbjct 60 YLDTINRKLLDFDFEKVCSVSLTNLSVYACLVCGRYFQGRGPSSHAYFHALTENHHVFVN 119
Query 143 LTTCKVYCL 151
+T K Y L
Sbjct 120 CSTLKFYVL 128
> At4g22410
Length=340
Score = 87.8 bits (216), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 38/61 (62%), Positives = 48/61 (78%), Gaps = 0/61 (0%)
Query 91 MLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMHALEQRHFVYVNLTTCKVYC 150
+LDF FE+ CS+ LSN +VYACLVCG+YFQGR + + A+ H+LE H VY+NL T KVYC
Sbjct 6 VLDFHFERFCSVSLSNLNVYACLVCGKYFQGRSQKSHAYTHSLEAGHHVYINLLTEKVYC 65
Query 151 L 151
L
Sbjct 66 L 66
> YFR005c
Length=448
Score = 81.6 bits (200), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 37/80 (46%), Positives = 54/80 (67%), Gaps = 3/80 (3%)
Query 75 KRVRRTCP---YLGTINKHMLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMH 131
K+++ P YL T+ + LDFD EK+C I LS +VY CLVCG Y+QGR + AF+H
Sbjct 20 KKIKSQEPNYAYLETVVREKLDFDSEKICCITLSPLNVYCCLVCGHYYQGRHEKSPAFIH 79
Query 132 ALEQRHFVYVNLTTCKVYCL 151
++++ H V++NLT+ K Y L
Sbjct 80 SIDENHHVFLNLTSLKFYML 99
> CE25887
Length=955
Score = 35.8 bits (81), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 32/72 (44%), Gaps = 1/72 (1%)
Query 80 TCPYLGTINK-HMLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMHALEQRHF 138
TCP+L + + CS C V+ CL C +Y GR + A MH L H
Sbjct 854 TCPHLKEVKPLPPAKINARTACSECQIGAEVWTCLTCYKYNCGRFVNEHAMMHHLSSSHP 913
Query 139 VYVNLTTCKVYC 150
+ +++ V+C
Sbjct 914 MALSMADLSVWC 925
> SPAC16E8.13
Length=547
Score = 35.0 bits (79), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 15/55 (27%), Positives = 29/55 (52%), Gaps = 0/55 (0%)
Query 95 DFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMHALEQRHFVYVNLTTCKVY 149
+F+ C++C ++ ++ CL+CG GR A H ++ H + L T +V+
Sbjct 257 EFQSKCTVCCYDKDLWICLICGNIGCGRYHDAHAKQHYVDTAHCYAMELETQRVW 311
> 7293562
Length=481
Score = 27.7 bits (60), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 32 EDSSSNDSSSSATNNSSTTSDNTNSNTSSSNNGSSSSSSAP 72
E SS+ D +SSA N SS + T+S +++ +SSAP
Sbjct 209 ESSSAPDQTSSAPNESSAPPEETSSAPEQTSSAPEETSSAP 249
Lambda K H
0.311 0.119 0.337
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1888713112
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40