bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1267_orf1
Length=138
Score E
Sequences producing significant alignments: (Bits) Value
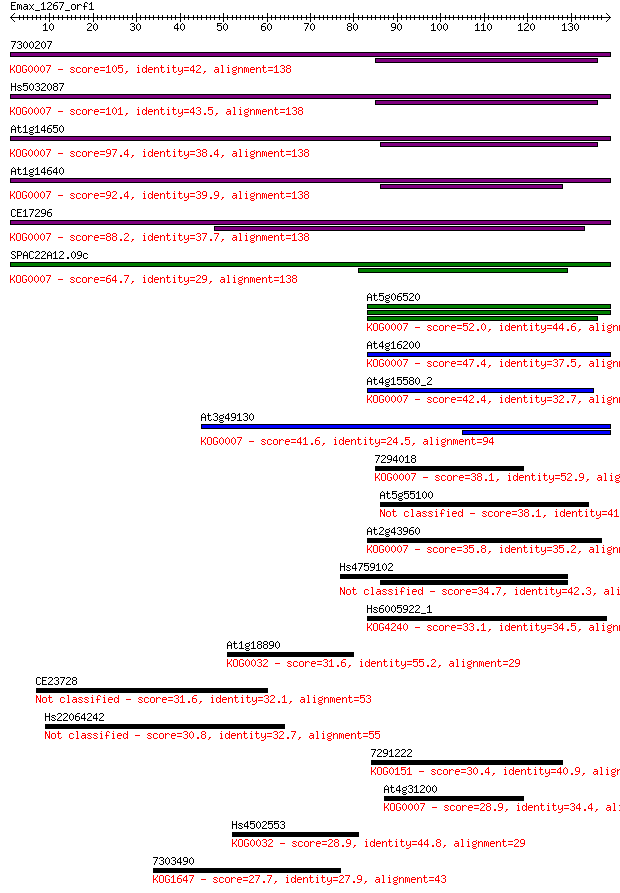
7300207 105 4e-23
Hs5032087 101 3e-22
At1g14650 97.4 7e-21
At1g14640 92.4 3e-19
CE17296 88.2 4e-18
SPAC22A12.09c 64.7 6e-11
At5g06520 52.0 3e-07
At4g16200 47.4 8e-06
At4g15580_2 42.4 3e-04
At3g49130 41.6 5e-04
7294018 38.1 0.005
At5g55100 38.1 0.005
At2g43960 35.8 0.024
Hs4759102 34.7 0.057
Hs6005922_1 33.1 0.15
At1g18890 31.6 0.41
CE23728 31.6 0.48
Hs22064242 30.8 0.72
7291222 30.4 0.92
At4g31200 28.9 2.9
Hs4502553 28.9 3.0
7303490 27.7 6.0
> 7300207
Length=784
Score = 105 bits (261), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 58/141 (41%), Positives = 83/141 (58%), Gaps = 14/141 (9%)
Query 1 NPYHAFYKLKVREFTTGEAAPTPQVPQAIRDMQQKQQQQKQKQQQLLMLTQIDESEKDGV 60
+PYHA+Y+ KV EF G A A+ M+Q +Q+Q +L Q+ V
Sbjct 74 DPYHAYYRHKVNEFREGNDAGI----TALASMKQLAVTSAAQQRQQELLKQV-------V 122
Query 61 KEKL---EPPPPNQFVLQHPWVAPVDLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAF 117
+++ EPPP +F+ P ++ +DLDI++ AQFVARNG++FL L REQ+N QF F
Sbjct 123 EQQFVPKEPPPEFEFIADPPSISALDLDIVKLTAQFVARNGRQFLTNLMSREQRNFQFDF 182
Query 118 LKPSHHLFSYFASLVDAYTKC 138
L+P H LF YF L++ YTK
Sbjct 183 LRPQHSLFQYFTKLLEQYTKV 203
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 28/51 (54%), Gaps = 0/51 (0%)
Query 85 DIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDAY 135
+I+ A FVARNG +F A + Q E NP+F FL +Y+ V+ +
Sbjct 37 NIVDKTASFVARNGPEFEARIRQNELGNPKFNFLNGGDPYHAYYRHKVNEF 87
> Hs5032087
Length=793
Score = 101 bits (252), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 60/141 (42%), Positives = 89/141 (63%), Gaps = 13/141 (9%)
Query 1 NPYHAFYKLKVREFTTGEA-APTPQVPQAIRDMQQKQQQQ--KQKQQQLLMLTQIDESEK 57
+PYHA+Y+ KV EF G+A P+ +P+ ++ QQ QQQ ++ Q Q++ T + +
Sbjct 88 DPYHAYYRHKVSEFKEGKAQEPSAAIPKVMQQQQQTTQQQLPQKVQAQVIQETIVPK--- 144
Query 58 DGVKEKLEPPPPNQFVLQHPWVAPVDLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAF 117
EPPP +F+ P ++ DLD+++ AQFVARNG++FL L Q+EQ+N QF F
Sbjct 145 -------EPPPEFEFIADPPSISAFDLDVVKLTAQFVARNGRQFLTQLMQKEQRNYQFDF 197
Query 118 LKPSHHLFSYFASLVDAYTKC 138
L+P H LF+YF LV+ YTK
Sbjct 198 LRPQHSLFNYFTKLVEQYTKI 218
Score = 43.9 bits (102), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 29/51 (56%), Gaps = 0/51 (0%)
Query 85 DIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDAY 135
+I+ A FVARNG +F A + Q E NP+F FL P+ +Y+ V +
Sbjct 51 NIVDKTASFVARNGPEFEARIRQNEINNPKFNFLNPNDPYHAYYRHKVSEF 101
> At1g14650
Length=785
Score = 97.4 bits (241), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 53/150 (35%), Positives = 79/150 (52%), Gaps = 23/150 (15%)
Query 1 NPYHAFYKLKVREFTT-----------GEAAPTPQVPQAIRDMQQKQQQQKQKQQQLLML 49
+PYHAFY+ K+ E+ + PQ+ D + Q Q Q +
Sbjct 107 DPYHAFYQHKLTEYRAQNKDGAQGTDDSDGTTDPQLDTGAADESEAGDTQPDLQAQFRIP 166
Query 50 TQIDESEKDGVKEKLEPPPPNQFVLQHP-WVAPVDLDIIRCCAQFVARNGQKFLAGLAQR 108
++ LE P P ++ ++ P + +LDII+ AQFVARNG+ FL GL+ R
Sbjct 167 SK-----------PLEAPEPEKYTVRLPEGITGEELDIIKLTAQFVARNGKSFLTGLSNR 215
Query 109 EQQNPQFAFLKPSHHLFSYFASLVDAYTKC 138
E NPQF F+KP+H +F++F SLVDAY++
Sbjct 216 ENNNPQFHFMKPTHSMFTFFTSLVDAYSEV 245
Score = 32.7 bits (73), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 15/50 (30%), Positives = 27/50 (54%), Gaps = 0/50 (0%)
Query 86 IIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDAY 135
I+ AQFV++NG +F + ++N +F FLK S +++ + Y
Sbjct 71 IVEKTAQFVSKNGLEFEKRIIVSNEKNAKFNFLKSSDPYHAFYQHKLTEY 120
> At1g14640
Length=735
Score = 92.4 bits (228), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 55/147 (37%), Positives = 78/147 (53%), Gaps = 25/147 (17%)
Query 1 NPYHAFYKLKVREFTTGEAAPTPQVPQAIRDMQQKQQQQKQKQQQLLMLTQIDESEKD-- 58
NPYH FY+ KV E++ IRD Q + +L DES+
Sbjct 106 NPYHGFYRYKVTEYSC-----------HIRDGAQGTDVDDTEDPKL-----DDESDAKPD 149
Query 59 ------GVKEKLEPPPPNQFVLQHP-WVAPVDLDIIRCCAQFVARNGQKFLAGLAQREQQ 111
++ LE P P ++ ++ P + +LDII+ AQFVARNGQ FL L +RE
Sbjct 150 LQAQFRAPRKILEAPEPEKYTVRLPEGIMEAELDIIKHTAQFVARNGQSFLRELMRREVN 209
Query 112 NPQFAFLKPSHHLFSYFASLVDAYTKC 138
N QF F+KP+H +F++F SLVDAY++
Sbjct 210 NSQFQFMKPTHSMFTFFTSLVDAYSEV 236
Score = 33.5 bits (75), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 23/45 (51%), Gaps = 3/45 (6%)
Query 86 IIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLK---PSHHLFSY 127
I+ AQFV++NG F + + N F+FLK P H + Y
Sbjct 70 IVETTAQFVSQNGLAFGNKVKTEKANNANFSFLKSDNPYHGFYRY 114
> CE17296
Length=655
Score = 88.2 bits (217), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 52/138 (37%), Positives = 70/138 (50%), Gaps = 24/138 (17%)
Query 1 NPYHAFYKLKVREFTTGEAAPTPQVPQAIRDMQQKQQQQKQKQQQLLMLTQIDESEKDGV 60
+PYHA+YK V +F+ G P+VPQA+++ +K +
Sbjct 73 DPYHAYYKKMVYDFSEGRVE-APKVPQAVKEHVKKAEFVPSA------------------ 113
Query 61 KEKLEPPPPNQFVLQHPWVAPVDLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKP 120
PPP +F + DLD+IR A FVARNG++FL L RE +N QF FLKP
Sbjct 114 -----PPPAYEFSADPSTINAYDLDLIRLVALFVARNGRQFLTQLMTREARNYQFDFLKP 168
Query 121 SHHLFSYFASLVDAYTKC 138
+H F+YF LVD Y K
Sbjct 169 AHCNFTYFTKLVDQYQKV 186
Score = 37.0 bits (84), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 21/85 (24%), Positives = 40/85 (47%), Gaps = 2/85 (2%)
Query 48 MLTQIDESEKDGVKEKLEPPPPNQFVLQHPWVAPVDLDIIRCCAQFVARNGQKFLAGLAQ 107
M + E+D + EP + ++ + P I+ A+F A+NG F + +
Sbjct 1 MTAVVSNREEDSMNN--EPSLSGRAIIGLIYPPPDIRTIVDKTARFAAKNGVDFENKIRE 58
Query 108 REQQNPQFAFLKPSHHLFSYFASLV 132
+E +NP+F FL + +Y+ +V
Sbjct 59 KEAKNPKFNFLSITDPYHAYYKKMV 83
> SPAC22A12.09c
Length=481
Score = 64.7 bits (156), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 40/142 (28%), Positives = 66/142 (46%), Gaps = 26/142 (18%)
Query 1 NPYHAFYKLKVREFTTGEAAPTPQVPQAIRDMQQKQQQQKQKQQQLLMLTQIDESEKDGV 60
+PYH +Y+ K+ E G K K + TQ + +
Sbjct 80 DPYHPYYQHKLTEAREG----------------------KLKSHATGLSTQKTSTLARPI 117
Query 61 KEKLEP--PPPNQFVLQHPW--VAPVDLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFA 116
++ +E P P+ ++ P ++ +DLD++R A++ A G FL L+Q+E N QF
Sbjct 118 QKPIEATIPAPSPYLFSEPLPSISSLDLDVLRLTARYAAVRGSSFLVSLSQKEWNNTQFD 177
Query 117 FLKPSHHLFSYFASLVDAYTKC 138
FLKP++ L+ YF +V YT
Sbjct 178 FLKPNNALYPYFMRIVQQYTSL 199
Score = 37.4 bits (85), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 19/48 (39%), Positives = 25/48 (52%), Gaps = 0/48 (0%)
Query 81 PVDLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYF 128
P +II A +VARNG F + Q EQ N +FAFL + Y+
Sbjct 39 PAIREIIDKSASYVARNGPAFEEKIRQNEQANTKFAFLHANDPYHPYY 86
> At5g06520
Length=679
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 25/57 (43%), Positives = 38/57 (66%), Gaps = 1/57 (1%)
Query 83 DLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHH-LFSYFASLVDAYTKC 138
+L II+ AQF+AR G F+ GL +R NPQF FL+ +++ FS++ LV AY++
Sbjct 487 ELGIIKLTAQFMARYGMNFVQGLRKRVVGNPQFKFLESTNNSRFSFYNGLVIAYSRV 543
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 31/56 (55%), Gaps = 0/56 (0%)
Query 83 DLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDAYTKC 138
+LD I+ AQFVA G F L +R +P+F F K + S++ LVD Y++
Sbjct 218 ELDTIKLTAQFVAVYGTLFRTELMKRVFISPKFDFFKSTDSKCSFYLRLVDGYSRV 273
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 31/53 (58%), Gaps = 0/53 (0%)
Query 83 DLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDAY 135
+L++I+ AQFVA G+ F L R ++P F FLKP+ S++ ++ Y
Sbjct 89 ELELIKLTAQFVAVYGKYFQRELTTRVVESPLFEFLKPTDSRNSFYTRIILGY 141
> At4g16200
Length=288
Score = 47.4 bits (111), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 32/56 (57%), Gaps = 0/56 (0%)
Query 83 DLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDAYTKC 138
+LD ++ AQFVA G F L +R PQF F+K + + FS++ V AY++
Sbjct 200 ELDTVKVTAQFVAWYGDDFRGFLMERVMTEPQFEFMKATDYRFSFYNEFVVAYSQV 255
> At4g15580_2
Length=375
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 31/52 (59%), Gaps = 0/52 (0%)
Query 83 DLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDA 134
+LD ++ AQFVA G F L +R + QF F+K + ++F++F + ++
Sbjct 183 ELDTVKVTAQFVAWYGDAFRGKLMERVMMDNQFEFMKQTDYMFAFFTAYMNT 234
> At3g49130
Length=307
Score = 41.6 bits (96), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 23/94 (24%), Positives = 45/94 (47%), Gaps = 3/94 (3%)
Query 45 QLLMLTQIDESEKDGVKEKLEPPPPNQFVLQHPWVAPVDLDIIRCCAQFVARNGQKFLAG 104
++++ ++ +++ D V L+ P +Q + + +L +++ AQ R G F G
Sbjct 24 SVMIMNRVAQTQPDMV---LQLPLGSQLTIPAKGITLKELGVMKLTAQSSVRYGFDFWCG 80
Query 105 LAQREQQNPQFAFLKPSHHLFSYFASLVDAYTKC 138
L +R NP F FL PS ++ AY++
Sbjct 81 LWKRVYMNPLFQFLNPSDSRSDFYNGFTAAYSRV 114
Score = 31.2 bits (69), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 11/34 (32%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 105 LAQREQQNPQFAFLKPSHHLFSYFASLVDAYTKC 138
L ++ PQF F++P++ +F + +VDAY++
Sbjct 226 LMKKVVMKPQFKFMEPTNSVFGLYNVVVDAYSRV 259
> 7294018
Length=182
Score = 38.1 bits (87), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 18/34 (52%), Positives = 24/34 (70%), Gaps = 0/34 (0%)
Query 85 DIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFL 118
+II A+FVARNG +F A Q++Q NP+F FL
Sbjct 13 NIIDKLAEFVARNGPEFEAITKQKQQNNPKFEFL 46
> At5g55100
Length=875
Score = 38.1 bits (87), Expect = 0.005, Method: Composition-based stats.
Identities = 20/48 (41%), Positives = 27/48 (56%), Gaps = 0/48 (0%)
Query 86 IIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVD 133
II + FV+++G + L ++ NP F FL P HHL YF LVD
Sbjct 184 IITRTSSFVSKHGGQSEIVLRVKQGDNPTFGFLMPDHHLHLYFRFLVD 231
> At2g43960
Length=421
Score = 35.8 bits (81), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 26/54 (48%), Gaps = 0/54 (0%)
Query 83 DLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDAYT 136
+L II+ AQFV R GQ F L R + +F FL +F+ L Y+
Sbjct 104 ELGIIKFTAQFVLRYGQYFRLALRDRVSTDTEFKFLDKGDSRAHFFSLLFLGYS 157
> Hs4759102
Length=951
Score = 34.7 bits (78), Expect = 0.057, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 30/67 (44%), Gaps = 15/67 (22%)
Query 77 PWVAPVDLD---------------IIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPS 121
P+VAP+ L II A FV R G +F L ++ N QF FL+
Sbjct 187 PFVAPLGLSVPSDVELPPTAKMHAIIERTASFVCRQGAQFEIMLKAKQAPNSQFDFLRFD 246
Query 122 HHLFSYF 128
H+L Y+
Sbjct 247 HYLNPYY 253
Score = 31.6 bits (70), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 26/43 (60%), Gaps = 2/43 (4%)
Query 86 IIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYF 128
+I A++VARNG KF + R + + +F FL+P + +Y+
Sbjct 459 VIDKLAEYVARNGLKFETSV--RAKNDQRFEFLQPWYQYNAYY 499
> Hs6005922_1
Length=2559
Score = 33.1 bits (74), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 29/56 (51%), Gaps = 1/56 (1%)
Query 83 DLDIIRCCAQFVARNGQKFLAGLAQR-EQQNPQFAFLKPSHHLFSYFASLVDAYTK 137
D+D+IR CA+ VA + Q+ + + R + N AF K S + S SL Y +
Sbjct 922 DMDMIRDCAEKVASHWQQLMLKMEDRLKLVNASVAFYKTSEQVCSVLESLEQEYKR 977
> At1g18890
Length=545
Score = 31.6 bits (70), Expect = 0.41, Method: Composition-based stats.
Identities = 16/33 (48%), Positives = 19/33 (57%), Gaps = 4/33 (12%)
Query 51 QIDESEKDGVKEKLEPPPPN----QFVLQHPWV 79
QI ES K VK+ L+P P Q VL HPW+
Sbjct 289 QISESAKSLVKQMLDPDPTKRLTAQQVLAHPWI 321
> CE23728
Length=493
Score = 31.6 bits (70), Expect = 0.48, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 30/53 (56%), Gaps = 2/53 (3%)
Query 7 YKLKVREFTTGEAAPTPQVPQAIRDMQQKQQQQKQKQQQLLMLTQIDESEKDG 59
Y + E+ AAP A+R ++KQ++ +++QQ ++ TQ E +KDG
Sbjct 356 YSIVTDEYVVLPAAPVKTF--AVRIQEEKQKKLDEEEQQKVLKTQELEKQKDG 406
> Hs22064242
Length=486
Score = 30.8 bits (68), Expect = 0.72, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 32/55 (58%), Gaps = 1/55 (1%)
Query 9 LKVREFTTGEAAPTPQVPQAIRDMQQKQQQQKQKQQQLLMLTQIDESEKDGVKEK 63
LK T+GE P P +R+M+QK QQ+++ +Q L L ++ ++E+ V +
Sbjct 415 LKKLRQTSGEVGLAPTDP-VLREMEQKLQQEEEDRQLALQLQRMFDNERRTVSRR 468
> 7291222
Length=957
Score = 30.4 bits (67), Expect = 0.92, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 26/48 (54%), Gaps = 4/48 (8%)
Query 84 LDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFL----KPSHHLFSY 127
L++I +FV R G F A + RE +NP FAFL P+H + +
Sbjct 372 LNVIHRMIEFVIREGPMFEALIMIREMENPLFAFLFDNESPAHIYYRW 419
> At4g31200
Length=650
Score = 28.9 bits (63), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 11/32 (34%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 87 IRCCAQFVARNGQKFLAGLAQREQQNPQFAFL 118
I ++ +NG +F A + R++ NP +AFL
Sbjct 130 IDKLVEYSVKNGPEFEAMMRDRQKDNPDYAFL 161
> Hs4502553
Length=370
Score = 28.9 bits (63), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 19/33 (57%), Gaps = 4/33 (12%)
Query 52 IDESEKDGVKEKLEPPPPNQFV----LQHPWVA 80
I +S KD ++ +E P +F LQHPW+A
Sbjct 245 ISDSAKDFIRHLMEKDPEKRFTCEQALQHPWIA 277
> 7303490
Length=373
Score = 27.7 bits (60), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 12/43 (27%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 34 QKQQQQKQKQQQLLMLTQIDESEKDGVKEKLEPPPPNQFVLQH 76
++++ +++ ++ L L Q E E+ ++ EPPPP F+ H
Sbjct 256 EEREVKRRMREARLSLKQRREQERKEAIKRGEPPPPTSFITTH 298
Lambda K H
0.320 0.134 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1498437086
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40