bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1253_orf1
Length=87
Score E
Sequences producing significant alignments: (Bits) Value
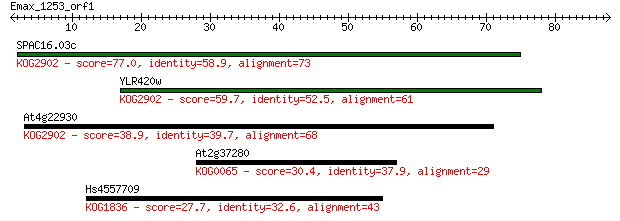
SPAC16.03c 77.0 9e-15
YLR420w 59.7 1e-09
At4g22930 38.9 0.002
At2g37280 30.4 0.79
Hs4557709 27.7 4.9
> SPAC16.03c
Length=337
Score = 77.0 bits (188), Expect = 9e-15, Method: Composition-based stats.
Identities = 43/77 (55%), Positives = 50/77 (64%), Gaps = 4/77 (5%)
Query 2 GLTLHIHAE---SRKATALKAEETFLPAFEQIHQRFPGLKIVFEHISTAAAVEAVKG-KK 57
G+ L+IH E S+ T AE FLP +HQRFP LKIV EH +TA AVEAVK +
Sbjct 126 GMILNIHGEVPPSKDNTVFTAEPKFLPTLLDLHQRFPKLKIVLEHCTTADAVEAVKACGE 185
Query 58 NVAATITAHHLRLTTGD 74
+VA TITAHHL LT D
Sbjct 186 SVAGTITAHHLYLTQKD 202
> YLR420w
Length=364
Score = 59.7 bits (143), Expect = 1e-09, Method: Composition-based stats.
Identities = 32/70 (45%), Positives = 44/70 (62%), Gaps = 9/70 (12%)
Query 17 LKAEETFLPAFEQIHQRFPGLKIVFEHISTAAAVEAVKG-KKN--------VAATITAHH 67
L AEE FLPA +++H FP LKI+ EH ++ +A++ ++ KN VAAT+TAHH
Sbjct 154 LNAEEAFLPALKKLHNDFPNLKIILEHCTSESAIKTIEDINKNVKKATDVKVAATLTAHH 213
Query 68 LRLTTGDVWG 77
L LT D G
Sbjct 214 LFLTIDDWAG 223
> At4g22930
Length=377
Score = 38.9 bits (89), Expect = 0.002, Method: Composition-based stats.
Identities = 27/73 (36%), Positives = 38/73 (52%), Gaps = 5/73 (6%)
Query 3 LTLHIHAESRKAT--ALKAEETFLPAFEQ-IHQRFPGLKIVFEHISTAAAVEAVKGKK-- 57
+ L +H E + E+ F+ Q + QR P LK+V EHI+T AV V+ K
Sbjct 163 MPLLVHGEVTDPSIDVFDREKIFIETVLQPLIQRLPQLKVVMEHITTMDAVNFVESCKEG 222
Query 58 NVAATITAHHLRL 70
+V AT+T HL L
Sbjct 223 SVGATVTPQHLLL 235
> At2g37280
Length=1413
Score = 30.4 bits (67), Expect = 0.79, Method: Composition-based stats.
Identities = 11/29 (37%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 28 EQIHQRFPGLKIVFEHISTAAAVEAVKGK 56
E++ FP +++ +EH+ AA E V+GK
Sbjct 95 ERVGVEFPSIEVRYEHLGVEAACEVVEGK 123
> Hs4557709
Length=3110
Score = 27.7 bits (60), Expect = 4.9, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 12 RKATALKAEETFLPAFEQIHQRFPGLKIVFEHISTAAAVEAVK 54
R LKAE+T LP ++ Q IVF+H A ++ ++
Sbjct 1183 RTWVTLKAEQTILPLVDEALQHTTTKGIVFQHPEIVAHMDLMR 1225
Lambda K H
0.317 0.129 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187582654
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40