bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1203_orf1
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
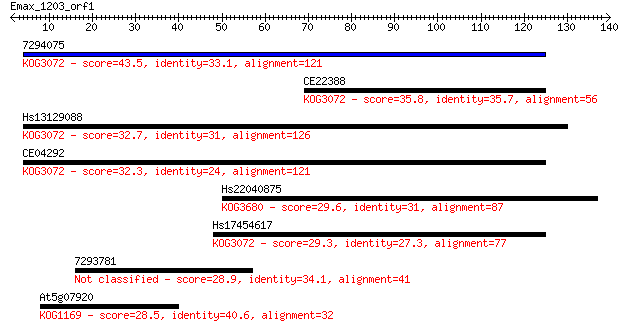
7294075 43.5 1e-04
CE22388 35.8 0.027
Hs13129088 32.7 0.20
CE04292 32.3 0.29
Hs22040875 29.6 2.0
Hs17454617 29.3 2.6
7293781 28.9 3.4
At5g07920 28.5 3.9
> 7294075
Length=313
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 40/134 (29%), Positives = 62/134 (46%), Gaps = 30/134 (22%)
Query 4 WLLLLPVLPHMWYMGSSSAASGLWVCVCLEDCVGPSSYRQPTTCACLLSRQALSAAAAAL 63
W + VL + W+ + +S W V + CV + + S AL AA
Sbjct 145 WYHHITVLIYSWFSYTEYTSSARWFIV-MNYCVH----------SVMYSYYALKAA---- 189
Query 64 RQSRPA-----ITLMQIAQMLVG--------MYLATKGALCLHDQQQVLNAQIALAMYTS 110
R + P IT +Q+AQM++G +L T G H Q+ +N +++AMY+S
Sbjct 190 RFNPPRFISMIITSLQLAQMIIGCAINVWANGFLKTHGTSSCHISQRNIN--LSIAMYSS 247
Query 111 YAVLFLRLYCSSYL 124
Y VLF R + +YL
Sbjct 248 YFVLFARFFYKAYL 261
> CE22388
Length=286
Score = 35.8 bits (81), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 32/63 (50%), Gaps = 10/63 (15%)
Query 69 AITLMQIAQMLVG-------MYLATKGALCLHDQQQVLNAQIALAMYTSYAVLFLRLYCS 121
+T +Q QML+G +YL G +C QQ N ++ +Y S+ VLF + +
Sbjct 215 TVTFLQTLQMLIGVSISCIVLYLKLNGEMC---QQSYDNLALSFGIYASFLVLFSSFFNN 271
Query 122 SYL 124
+YL
Sbjct 272 AYL 274
> Hs13129088
Length=265
Score = 32.7 bits (73), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 39/133 (29%), Positives = 49/133 (36%), Gaps = 21/133 (15%)
Query 4 WLLLLPVLPHMWYMGSSSAASGLWVCVCLEDCVGPSSYRQPTTCACLLSRQALSAAAAAL 63
W + VL + WY A G W A + S AL AA
Sbjct 142 WYHHITVLLYSWYSYKDMVAGGGWFMTM-----------NYGVHAVMYSYYALRAAG--F 188
Query 64 RQSRPA---ITLMQIAQMLVGMYLATKGALCL--HDQ--QQVLNAQIALAMYTSYAVLFL 116
R SR ITL QI QML+G + C HDQ N + MY SY VLF
Sbjct 189 RVSRKFAMFITLSQITQMLMGC-VVNYLVFCWMQHDQCHSHFQNIFWSSLMYLSYLVLFC 247
Query 117 RLYCSSYLPHLSS 129
+ +Y+ +
Sbjct 248 HFFFEAYIGKMRK 260
> CE04292
Length=435
Score = 32.3 bits (72), Expect = 0.29, Method: Composition-based stats.
Identities = 29/129 (22%), Positives = 52/129 (40%), Gaps = 22/129 (17%)
Query 4 WLLLLPVLPHMWYMGSSSAASGLWVCVCLEDCVGPSSYRQPTTCACLLSRQALSAAAAAL 63
W + V+ + W+ ASG W + A + S AL + L
Sbjct 187 WYHHVTVMIYTWHAYKDHTASGRWFI-----------WMNYGVHALMYSYYALRSLKFRL 235
Query 64 -RQSRPAITLMQIAQMLVGMYLA-------TKGALCLHDQQQVLNAQIALAMYTSYAVLF 115
+Q +T +Q+AQM++G+ + + G C QQ N + +Y +Y +LF
Sbjct 236 PKQMAMVVTTLQLAQMVMGVIIGVTVYRIKSSGEYC---QQTWDNLGLCFGVYFTYFLLF 292
Query 116 LRLYCSSYL 124
+ +Y+
Sbjct 293 ANFFYHAYV 301
> Hs22040875
Length=742
Score = 29.6 bits (65), Expect = 2.0, Method: Composition-based stats.
Identities = 27/103 (26%), Positives = 49/103 (47%), Gaps = 16/103 (15%)
Query 50 LLSRQALSAAAAALRQSRPAITL---MQIAQMLVGMYL--ATKGALCLHDQ------QQV 98
LL+ + ++AA + +PAI+L + +L MYL T C D+ +Q
Sbjct 93 LLNMEIDNSAACSSAVRKPAISLADSTDLRVLLNIMYLIVETVHQECEGDKAEWRTMRQT 152
Query 99 LNAQIALAMYTS--YAVLFLRL---YCSSYLPHLSSNRVLLLF 136
A++ +Y + +A++ + +CS + PH +VLLL
Sbjct 153 FRAELGSPLYNNEPFAIMLFGMVTKFCSGHAPHFPMKKVLLLL 195
> Hs17454617
Length=270
Score = 29.3 bits (64), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 37/81 (45%), Gaps = 4/81 (4%)
Query 48 ACLLSRQALSAAAAALRQSRPA-ITLMQIAQMLVGMYLATKGALCLHDQ---QQVLNAQI 103
A + + L AA + P IT +QI QM VG ++ + DQ + +
Sbjct 179 AIMYTYYTLKAANVKPPKMLPMLITSLQILQMFVGAIVSILTYIWRQDQGCHTTMEHLFW 238
Query 104 ALAMYTSYAVLFLRLYCSSYL 124
+ +Y +Y +LF +C +Y+
Sbjct 239 SFILYMTYFILFAHFFCQTYI 259
> 7293781
Length=1339
Score = 28.9 bits (63), Expect = 3.4, Method: Composition-based stats.
Identities = 14/49 (28%), Positives = 26/49 (53%), Gaps = 8/49 (16%)
Query 16 YMGSSSAASGLWVCVCLEDCVGPSS--------YRQPTTCACLLSRQAL 56
Y+G ++ A+GLW V L++ +G ++ ++ PT C + Q L
Sbjct 26 YVGRTNFAAGLWYGVVLDEPLGKNNGSVHGSIYFKCPTNCGLFVRAQQL 74
> At5g07920
Length=728
Score = 28.5 bits (62), Expect = 3.9, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 18/32 (56%), Gaps = 1/32 (3%)
Query 8 LPVLPHMWYMGSSSAASGLWVCVCLEDCVGPS 39
+PV PH W + + A L CVCL+ + PS
Sbjct 75 VPVAPHSWELDPIARAKNLNCCVCLKS-MSPS 105
Lambda K H
0.328 0.134 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1534984332
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40