bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1192_orf2
Length=129
Score E
Sequences producing significant alignments: (Bits) Value
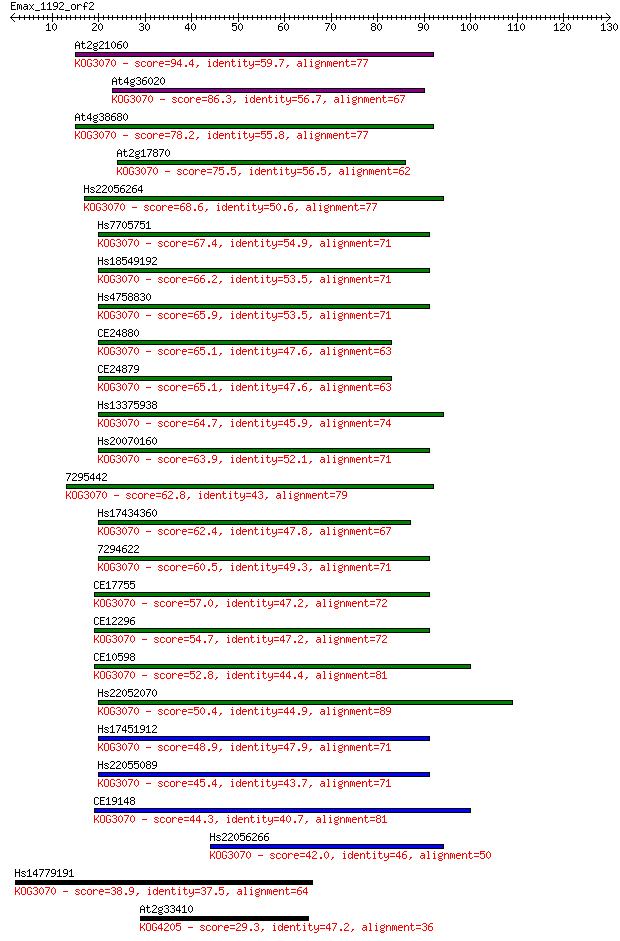
At2g21060 94.4 5e-20
At4g36020 86.3 1e-17
At4g38680 78.2 4e-15
At2g17870 75.5 3e-14
Hs22056264 68.6 3e-12
Hs7705751 67.4 6e-12
Hs18549192 66.2 1e-11
Hs4758830 65.9 2e-11
CE24880 65.1 3e-11
CE24879 65.1 3e-11
Hs13375938 64.7 4e-11
Hs20070160 63.9 7e-11
7295442 62.8 2e-10
Hs17434360 62.4 2e-10
7294622 60.5 7e-10
CE17755 57.0 8e-09
CE12296 54.7 5e-08
CE10598 52.8 2e-07
Hs22052070 50.4 7e-07
Hs17451912 48.9 2e-06
Hs22055089 45.4 2e-05
CE19148 44.3 6e-05
Hs22056266 42.0 3e-04
Hs14779191 38.9 0.002
At2g33410 29.3 2.0
> At2g21060
Length=201
Score = 94.4 bits (233), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 46/78 (58%), Positives = 60/78 (76%), Gaps = 1/78 (1%)
Query 15 GDIQRGTCKWFDSLKGFGFITSEDG-MDLFVHQTEIKAEGFRNLSEGEQVEFLVQTGSDG 73
GD ++GT KWFD+ KGFGFIT DG DLFVHQ+ I++EGFR+L+ E VEF V+ + G
Sbjct 12 GDRRKGTVKWFDTQKGFGFITPSDGGDDLFVHQSSIRSEGFRSLAAEESVEFDVEVDNSG 71
Query 74 RKKAVNVTGPGGAPVKGD 91
R KA+ V+GP GAPV+G+
Sbjct 72 RPKAIEVSGPDGAPVQGN 89
> At4g36020
Length=299
Score = 86.3 bits (212), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 38/68 (55%), Positives = 53/68 (77%), Gaps = 1/68 (1%)
Query 23 KWFDSLKGFGFITSEDG-MDLFVHQTEIKAEGFRNLSEGEQVEFLVQTGSDGRKKAVNVT 81
WF++ KG+GFIT +DG ++LFVHQ+ I +EG+R+L+ G+ VEF + GSDG+ KAVNVT
Sbjct 16 NWFNASKGYGFITPDDGSVELFVHQSSIVSEGYRSLTVGDAVEFAITQGSDGKTKAVNVT 75
Query 82 GPGGAPVK 89
PGG +K
Sbjct 76 APGGGSLK 83
> At4g38680
Length=203
Score = 78.2 bits (191), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 43/78 (55%), Positives = 62/78 (79%), Gaps = 1/78 (1%)
Query 15 GDIQRGTCKWFDSLKGFGFITSEDG-MDLFVHQTEIKAEGFRNLSEGEQVEFLVQTGSDG 73
G+ ++G+ KWFD+ KGFGFIT +DG DLFVHQ+ I++EGFR+L+ E VEF V+ ++
Sbjct 8 GERRKGSVKWFDTQKGFGFITPDDGGDDLFVHQSSIRSEGFRSLAAEEAVEFEVEIDNNN 67
Query 74 RKKAVNVTGPGGAPVKGD 91
R KA++V+GP GAPV+G+
Sbjct 68 RPKAIDVSGPDGAPVQGN 85
> At2g17870
Length=299
Score = 75.5 bits (184), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 35/63 (55%), Positives = 47/63 (74%), Gaps = 1/63 (1%)
Query 24 WFDSLKGFGFITSEDG-MDLFVHQTEIKAEGFRNLSEGEQVEFLVQTGSDGRKKAVNVTG 82
WF KG+GFIT +DG +LFVHQ+ I ++GFR+L+ GE VE+ + GSDG+ KA+ VT
Sbjct 15 WFSDGKGYGFITPDDGGEELFVHQSSIVSDGFRSLTLGESVEYEIALGSDGKTKAIEVTA 74
Query 83 PGG 85
PGG
Sbjct 75 PGG 77
> Hs22056264
Length=157
Score = 68.6 bits (166), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 39/87 (44%), Positives = 54/87 (62%), Gaps = 11/87 (12%)
Query 17 IQRGT--CKWFDSLKGFGFITS--------EDGMDLFVHQTEIKAEGFRNLSEGEQVEFL 66
+ RGT CKWF+ GFGFI+ + +D+FVHQ+++ EGFR+L EGE VEF
Sbjct 48 VLRGTGHCKWFNVRMGFGFISMINREGSPLDIPVDVFVHQSKLFMEGFRSLKEGEPVEFT 107
Query 67 VQTGSDGRKKAVNVTGPGGAPVKGDPR 93
+ S G +++ VTGPGG+P G R
Sbjct 108 FKKSSKGL-ESIRVTGPGGSPCLGSER 133
> Hs7705751
Length=364
Score = 67.4 bits (163), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 39/76 (51%), Positives = 46/76 (60%), Gaps = 6/76 (7%)
Query 20 GTCKWFDSLKGFGFITSEDGM-DLFVHQTEIKAEG----FRNLSEGEQVEFLVQTGSDGR 74
GT KWF+ G+GFI D D+FVHQT IK R++ +GE VEF V G G
Sbjct 96 GTVKWFNVRNGYGFINRNDTKEDVFVHQTAIKRNNPRKFLRSVGDGETVEFDVVEGEKG- 154
Query 75 KKAVNVTGPGGAPVKG 90
+A NVTGPGG PVKG
Sbjct 155 AEATNVTGPGGVPVKG 170
> Hs18549192
Length=324
Score = 66.2 bits (160), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 38/76 (50%), Positives = 46/76 (60%), Gaps = 6/76 (7%)
Query 20 GTCKWFDSLKGFGFITSEDGM-DLFVHQTEIKAEG----FRNLSEGEQVEFLVQTGSDGR 74
GT KWF+ G+GFI D D+FVHQT IK R++ +GE VEF V G G
Sbjct 61 GTVKWFNVRNGYGFINRNDTKEDVFVHQTAIKKNNPRKYLRSVGDGETVEFDVVEGEKG- 119
Query 75 KKAVNVTGPGGAPVKG 90
+A NVTGPGG PV+G
Sbjct 120 AEAANVTGPGGVPVQG 135
> Hs4758830
Length=322
Score = 65.9 bits (159), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 38/76 (50%), Positives = 46/76 (60%), Gaps = 6/76 (7%)
Query 20 GTCKWFDSLKGFGFITSEDGM-DLFVHQTEIKAEG----FRNLSEGEQVEFLVQTGSDGR 74
GT KWF+ G+GFI D D+FVHQT IK R++ +GE VEF V G G
Sbjct 60 GTVKWFNVRNGYGFINRNDTKEDVFVHQTAIKKNNPRKYLRSVGDGETVEFDVVEGEKG- 118
Query 75 KKAVNVTGPGGAPVKG 90
+A NVTGPGG PV+G
Sbjct 119 AEAANVTGPGGVPVQG 134
> CE24880
Length=196
Score = 65.1 bits (157), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 30/65 (46%), Positives = 47/65 (72%), Gaps = 2/65 (3%)
Query 20 GTCKWFDSLKGFGFITSE-DGMDLFVHQTEIKAEGFRNLSEGEQVEFLVQTGSDGR-KKA 77
G+CKWF+ KG+GF+ + G DLFVHQ+ + +GFR+L EGE+V + +Q S+G+ ++A
Sbjct 24 GSCKWFNVSKGYGFVIDDITGEDLFVHQSNLNMQGFRSLDEGERVSYYIQERSNGKGREA 83
Query 78 VNVTG 82
V+G
Sbjct 84 YAVSG 88
> CE24879
Length=227
Score = 65.1 bits (157), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 30/65 (46%), Positives = 47/65 (72%), Gaps = 2/65 (3%)
Query 20 GTCKWFDSLKGFGFITSE-DGMDLFVHQTEIKAEGFRNLSEGEQVEFLVQTGSDGR-KKA 77
G+CKWF+ KG+GF+ + G DLFVHQ+ + +GFR+L EGE+V + +Q S+G+ ++A
Sbjct 55 GSCKWFNVSKGYGFVIDDITGEDLFVHQSNLNMQGFRSLDEGERVSYYIQERSNGKGREA 114
Query 78 VNVTG 82
V+G
Sbjct 115 YAVSG 119
> Hs13375938
Length=209
Score = 64.7 bits (156), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 34/82 (41%), Positives = 49/82 (59%), Gaps = 9/82 (10%)
Query 20 GTCKWFDSLKGFGFITS--------EDGMDLFVHQTEIKAEGFRNLSEGEQVEFLVQTGS 71
G CKWF+ GFGF++ + +D+FVHQ+++ EGFR+L EGE VEF + +
Sbjct 42 GICKWFNVRMGFGFLSMTARAGVALDPPVDVFVHQSKLHMEGFRSLKEGEAVEFTFKKSA 101
Query 72 DGRKKAVNVTGPGGAPVKGDPR 93
G +++ VTGPGG G R
Sbjct 102 KGL-ESIRVTGPGGVFCIGSER 122
> Hs20070160
Length=372
Score = 63.9 bits (154), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 37/76 (48%), Positives = 45/76 (59%), Gaps = 6/76 (7%)
Query 20 GTCKWFDSLKGFGFITSEDGM-DLFVHQTEIKAEG----FRNLSEGEQVEFLVQTGSDGR 74
GT KWF+ G+GFI D D+FVHQT IK R++ +GE VEF V G G
Sbjct 93 GTVKWFNVRNGYGFINRNDTKEDVFVHQTAIKKNNPRKYLRSVGDGETVEFDVVEGEKG- 151
Query 75 KKAVNVTGPGGAPVKG 90
+A NVTGP G PV+G
Sbjct 152 AEAANVTGPDGVPVEG 167
> 7295442
Length=181
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 34/80 (42%), Positives = 49/80 (61%), Gaps = 2/80 (2%)
Query 13 KMGDIQRGTCKWFDSLKGFGFITSEDG-MDLFVHQTEIKAEGFRNLSEGEQVEFLVQTGS 71
+ G ++ G CKWF+ KG+GF+T DG ++FVHQ+ I+ GFR+L E E+VEF Q S
Sbjct 20 ECGCVRLGKCKWFNVAKGWGFLTPNDGGQEVFVHQSVIQMSGFRSLGEQEEVEFECQRTS 79
Query 72 DGRKKAVNVTGPGGAPVKGD 91
G +A V+ G +G
Sbjct 80 RGL-EATRVSSRHGGSCQGS 98
> Hs17434360
Length=181
Score = 62.4 bits (150), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/75 (42%), Positives = 47/75 (62%), Gaps = 9/75 (12%)
Query 20 GTCKWFDSLKGFGFITS--------EDGMDLFVHQTEIKAEGFRNLSEGEQVEFLVQTGS 71
G CKWF+ GFG ++ + +D+FVHQ+++ EGFR+L EGE VEF + +
Sbjct 101 GICKWFNLRMGFGLLSVTARAVVALDPPVDVFVHQSKLPMEGFRSLKEGEAVEFTFKKSA 160
Query 72 DGRKKAVNVTGPGGA 86
G +++ VTGPGGA
Sbjct 161 KG-LESIRVTGPGGA 174
> 7294622
Length=340
Score = 60.5 bits (145), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 35/76 (46%), Positives = 45/76 (59%), Gaps = 6/76 (7%)
Query 20 GTCKWFDSLKGFGFITSEDGM-DLFVHQTEI----KAEGFRNLSEGEQVEFLVQTGSDGR 74
GT KWF+ G+GFI D D+FVHQ+ I + R++ +GE VEF V G G
Sbjct 64 GTVKWFNVKSGYGFINRNDTREDVFVHQSAIARNNPKKAVRSVGDGEVVEFDVVIGEKG- 122
Query 75 KKAVNVTGPGGAPVKG 90
+A NVTGP G PV+G
Sbjct 123 NEAANVTGPSGEPVRG 138
> CE17755
Length=208
Score = 57.0 bits (136), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 34/77 (44%), Positives = 45/77 (58%), Gaps = 6/77 (7%)
Query 19 RGTCKWFDSLKGFGFITSED-GMDLFVHQTEIKAEG----FRNLSEGEQVEFLVQTGSDG 73
+GT KWF+ G+GFI D D+FVHQT I R+L + E+V F + GS G
Sbjct 22 KGTVKWFNVKNGYGFINRTDTNEDIFVHQTAIINNNPNKYLRSLGDNEEVMFDIVEGSKG 81
Query 74 RKKAVNVTGPGGAPVKG 90
+ A +VTGP G PV+G
Sbjct 82 LE-AASVTGPDGGPVQG 97
> CE12296
Length=265
Score = 54.7 bits (130), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 34/78 (43%), Positives = 46/78 (58%), Gaps = 7/78 (8%)
Query 19 RGTCKWFDSLKGFGFITSEDG-MDLFVHQTEIK---AEGF--RNLSEGEQVEFLVQTGSD 72
+G KW+ L+ +GFI+ DG D+FVHQT I E F R L++ E+V F + G +
Sbjct 65 QGKVKWYSVLRRYGFISRSDGEKDVFVHQTAISKSDTEKFYLRTLADEEEVLFDLVDGKN 124
Query 73 GRKKAVNVTGPGGAPVKG 90
G +A NVTGP G V G
Sbjct 125 G-PEAANVTGPAGVNVSG 141
> CE10598
Length=267
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 36/87 (41%), Positives = 47/87 (54%), Gaps = 8/87 (9%)
Query 19 RGTCKWFDSLKGFGFITSEDG-MDLFVHQTEIKAEG-----FRNLSEGEQVEFLVQTGSD 72
+G KW+ L+ +GFI+ DG D+FVHQT I R L + E+V F + G +
Sbjct 67 QGKVKWYSVLRRYGFISRNDGEKDIFVHQTAIAKSATEKFYLRTLGDDEEVLFDLVEGKN 126
Query 73 GRKKAVNVTGPGGAPVKGDPRPVRKLL 99
G +A NVTGP G V G R KLL
Sbjct 127 G-PEAANVTGPNGDNVIG-SRYRHKLL 151
> Hs22052070
Length=393
Score = 50.4 bits (119), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 40/105 (38%), Positives = 51/105 (48%), Gaps = 17/105 (16%)
Query 20 GTCKWFDSLKGFGFITSEDGM-DLFVHQTEIKAEG----FRNLSEGEQVEFLVQTGSDGR 74
GT K F+ G+GF D D+FVH T +K R++ +GE VEF V G G
Sbjct 185 GTAKRFNVRSGYGFTNRNDAKEDVFVHWTAVKRNNPRKFLRSVRDGETVEFDVVEGEKG- 243
Query 75 KKAVNVTGP--GGAPVKGD---------PRPVRKLLPSGGGGRGG 108
+A NVTGP G P+KG P P+ +PS G GG
Sbjct 244 AEATNVTGPRAAGVPMKGTVMPPTDVVAPPPMVAEIPSRGTEPGG 288
> Hs17451912
Length=457
Score = 48.9 bits (115), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 34/73 (46%), Positives = 40/73 (54%), Gaps = 6/73 (8%)
Query 20 GTCKWFDSLKGFGFITSED-GMDLFVHQT-EIKAEGFRNLSEGEQVEFLVQTGSDGRKKA 77
GT KWF+ G+ FI D D FV QT + K RN GE VEF V G G +A
Sbjct 225 GTVKWFNLRNGYAFINRNDIKEDTFVPQTAKTKNNPRRN---GETVEFDVVEGEKG-AEA 280
Query 78 VNVTGPGGAPVKG 90
+V GPGG PV+G
Sbjct 281 AHVIGPGGVPVQG 293
> Hs22055089
Length=469
Score = 45.4 bits (106), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 31/74 (41%), Positives = 41/74 (55%), Gaps = 4/74 (5%)
Query 20 GTCKWFDSLKGFGFITSEDGM-DLFVHQTEIKAEGFRNLS--EGEQVEFLVQTGSDGRKK 76
GT KWF+ +GFI D D+FV QT I+ + L + E +EF V G G +
Sbjct 57 GTVKWFNVRNRYGFINRNDTKEDVFVPQTAIENNPRKYLPRVDRETMEFDVVEGEKG-AE 115
Query 77 AVNVTGPGGAPVKG 90
A +VTGPGG V+G
Sbjct 116 AAHVTGPGGVLVQG 129
> CE19148
Length=294
Score = 44.3 bits (103), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 33/90 (36%), Positives = 43/90 (47%), Gaps = 11/90 (12%)
Query 19 RGTCKWFDSLKGFGFIT----SEDGMDLFVHQTEIKAEG-----FRNLSEGEQVEFLVQT 69
+G KWF +GF+ +++ D FVHQT I R L + E V F +
Sbjct 90 KGHVKWFSVRGRYGFVARDKPTDENEDFFVHQTAITKSSTIKFYLRTLDDDEPVVFDIVE 149
Query 70 GSDGRKKAVNVTGPGGAPVKGDPRPVRKLL 99
G G +A NVTGP G V+G R R LL
Sbjct 150 GLKG-PEAANVTGPDGENVRG-SRFARTLL 177
> Hs22056266
Length=194
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/50 (46%), Positives = 33/50 (66%), Gaps = 1/50 (2%)
Query 44 VHQTEIKAEGFRNLSEGEQVEFLVQTGSDGRKKAVNVTGPGGAPVKGDPR 93
V ++++ EGFR+L EGE VEF + S G +++ VTGPGG+P G R
Sbjct 8 VSKSKLFMEGFRSLKEGEPVEFTFKKSSKGL-ESIRVTGPGGSPCLGSER 56
> Hs14779191
Length=153
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 34/65 (52%), Gaps = 3/65 (4%)
Query 2 RKRDSCEGGITKMGDIQRGTCKWFDSLKGFGFITSEDGM-DLFVHQTEIKAEGFRNLSEG 60
R R G + +G CK F +G GFIT E+G D+FVH ++I EG EG
Sbjct 53 RTRTYSATARASAGPVFKGVCKQFSRSQGHGFITPENGSEDIFVHVSDI--EGEYVPVEG 110
Query 61 EQVEF 65
++V +
Sbjct 111 DEVTY 115
> At2g33410
Length=404
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 26/38 (68%), Gaps = 9/38 (23%)
Query 29 KGFGFIT--SEDGMDLFVHQTEIKAEGFRNLSEGEQVE 64
+GFGF++ SED +DL +H+T F +L+ G+QVE
Sbjct 151 RGFGFVSFDSEDSVDLVLHKT------FHDLN-GKQVE 181
Lambda K H
0.314 0.143 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1209785478
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40