bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1184_orf1
Length=130
Score E
Sequences producing significant alignments: (Bits) Value
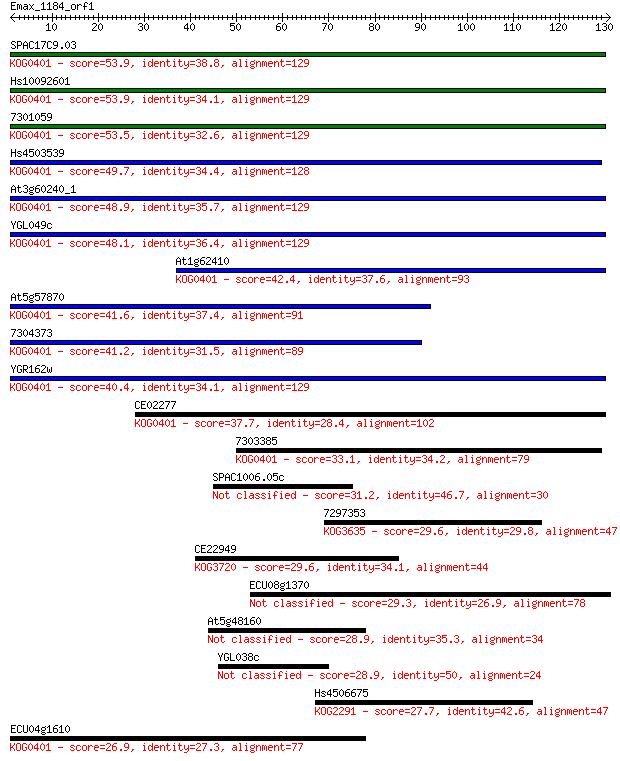
SPAC17C9.03 53.9 8e-08
Hs10092601 53.9 8e-08
7301059 53.5 9e-08
Hs4503539 49.7 1e-06
At3g60240_1 48.9 2e-06
YGL049c 48.1 4e-06
At1g62410 42.4 2e-04
At5g57870 41.6 4e-04
7304373 41.2 6e-04
YGR162w 40.4 7e-04
CE02277 37.7 0.005
7303385 33.1 0.15
SPAC1006.05c 31.2 0.46
7297353 29.6 1.3
CE22949 29.6 1.4
ECU08g1370 29.3 2.0
At5g48160 28.9 2.4
YGL038c 28.9 2.8
Hs4506675 27.7 5.1
ECU04g1610 26.9 9.9
> SPAC17C9.03
Length=1403
Score = 53.9 bits (128), Expect = 8e-08, Method: Composition-based stats.
Identities = 50/146 (34%), Positives = 75/146 (51%), Gaps = 20/146 (13%)
Query 1 LNKLTIEKFGVIAEKLAQILEKSLNCQAE-----IRIQVDAIIDKAVTEPDWSEMYADLC 55
LNK+T+EKF I++ QILE ++ + E ++ + +KA EP++S MYA
Sbjct 1016 LNKMTLEKFDKISD---QILEIAMQSRKENDGRTLKQVIQLTFEKATDEPNFSNMYARFA 1072
Query 56 QLLQWR---SVAPEG----DPETIRR-TPFMLGLLTRIQQEFE-AMPAALQHEVEGEAEE 106
+ + S+ EG + + +R F LL+R Q++FE A L GEAE
Sbjct 1073 RKMMDSIDDSIRDEGVLDKNNQPVRGGLLFRKYLLSRCQEDFERGWKANLPSGKAGEAEI 1132
Query 107 ALQE---EVRLKRRVLGVVKLIGELF 129
E +KRR LG+V+ IGELF
Sbjct 1133 MSDEYYVAAAIKRRGLGLVRFIGELF 1158
> Hs10092601
Length=1585
Score = 53.9 bits (128), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 44/145 (30%), Positives = 73/145 (50%), Gaps = 22/145 (15%)
Query 1 LNKLTIEKFGVIAEKLAQILEKSLNCQAEIRIQVDAIIDKAVTEPDWSEMYADLCQLLQW 60
LNKLT + F + ++++ + +++ + ++ +D + +KA+ EP +S YA++C+ L
Sbjct 762 LNKLTPQMFNQLMKQVSGL---TVDTEERLKGVIDLVFEKAIDEPSFSVAYANMCRCLVT 818
Query 61 RSVAPEGDP-ETIRRTPFMLGLLTRIQQEFEAMPAAL----QHEVEGEAEEALQEEVRL- 114
V P T+ F LL R Q+EFE A + + E EA A +E RL
Sbjct 819 LKVPMADKPGNTVN---FRKLLLNRCQKEFEKDKADDDVFEKKQKELEAASAPEERTRLH 875
Query 115 ----------KRRVLGVVKLIGELF 129
+RR +G +K IGELF
Sbjct 876 DELEEAKDKARRRSIGNIKFIGELF 900
> 7301059
Length=1905
Score = 53.5 bits (127), Expect = 9e-08, Method: Composition-based stats.
Identities = 42/148 (28%), Positives = 74/148 (50%), Gaps = 29/148 (19%)
Query 1 LNKLTIEKFGVIAEKLAQILEKSLNCQAEIRIQVDAIIDKAVTEPDWSEMYADLCQLLQW 60
LNKLT + F V+ ++++ I ++ +A++ + I +K ++EP+++ YA C++L +
Sbjct 1114 LNKLTPDNFEVLLKEMSSI---KMDNEAKMTNVMLLIFEKTISEPNFAPTYARFCKVL-F 1169
Query 61 RSVAPEGDPETIRRTPFMLGLLTRIQQEFE-------AMPAALQHEVE-----------G 102
+ E ++ F L+TRIQ EFE A LQ +E
Sbjct 1170 HEIKAEN------KSLFTSSLITRIQHEFESNVNDANAKSKKLQPTMERINQCSDPAKKA 1223
Query 103 EAEEALQE-EVRLKRRVLGVVKLIGELF 129
E +++ E + +RR G V+ IGELF
Sbjct 1224 ELRAEMEDLEYQFRRRAWGTVRFIGELF 1251
> Hs4503539
Length=907
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 44/137 (32%), Positives = 66/137 (48%), Gaps = 13/137 (9%)
Query 1 LNKLTIEKFGVIAEKLAQI-LEKSLNCQAEIRIQVDAIIDKAVTEPDWSEMYADLCQLL- 58
LNKLT EKF + +L + +E L + I + I+DKA+ EP +S +YA LC L
Sbjct 85 LNKLTPEKFDKLCLELLNVGVESKLILKGVILL----IVDKALEEPKYSSLYAQLCLRLA 140
Query 59 ----QWRSVAPEGDPETIRRTPFMLGLLTRIQQEFEAMPAAL---QHEVEGEAEEALQEE 111
+ A EG P + T F L++++Q EFE + E ++
Sbjct 141 EDAPNFDGPAAEGQPGQKQSTTFRRLLISKLQDEFENRTRNVDVYDKRENPLLPEEEEQR 200
Query 112 VRLKRRVLGVVKLIGEL 128
K ++LG +K IGEL
Sbjct 201 AIAKIKMLGNIKFIGEL 217
> At3g60240_1
Length=1528
Score = 48.9 bits (115), Expect = 2e-06, Method: Composition-based stats.
Identities = 46/139 (33%), Positives = 73/139 (52%), Gaps = 18/139 (12%)
Query 1 LNKLTIEKFGVIAEKLAQILEKSLNCQAEIRIQ--VDAIIDKAVTEPDWSEMYADLCQLL 58
LNKLT + F + E++ KS+N + + + I DKA+ EP + EMYAD C
Sbjct 920 LNKLTPQNFEKLFEQV-----KSVNIDNAVTLSGVISQIFDKALMEPTFCEMYADFC--F 972
Query 59 QWRSVAPEGDPETIRRTPFMLGLLTRIQQEFE--------AMPAALQHEVEGEAEEALQE 110
P+ + E + F LL + Q+EFE A A + +VE EE ++
Sbjct 973 HLSGALPDFN-ENGEKITFKRLLLNKCQEEFERGEKEEEEASRVAEEGQVEQTEEEREEK 1031
Query 111 EVRLKRRVLGVVKLIGELF 129
++++RR+LG ++LIGEL+
Sbjct 1032 RLQVRRRMLGNIRLIGELY 1050
> YGL049c
Length=914
Score = 48.1 bits (113), Expect = 4e-06, Method: Composition-based stats.
Identities = 47/153 (30%), Positives = 69/153 (45%), Gaps = 36/153 (23%)
Query 1 LNKLTIEKFGVIAEKLAQILEKSL--NCQAEIRIQVDAIIDKAVTEPDWSEMYADLCQLL 58
LNKLT+E F I+ ++ I +S + ++I ++ I KA EP WS MYA LC
Sbjct 574 LNKLTLEMFDSISSEILDIANQSKWEDDGETLKIVIEQIFHKACDEPHWSSMYAQLCG-- 631
Query 59 QWRSVAPEGDPETI------RRTPFML--GLLTRIQQEFE-----AMPAALQHEVEGEAE 105
V + DP + P ++ L+ R +EFE +PA GE
Sbjct 632 ---KVVKDLDPNIKDKENEGKNGPKLVLHYLVARCHEEFEKGWADKLPA-------GEDG 681
Query 106 EALQEEVR---------LKRRVLGVVKLIGELF 129
L+ E+ KRR LG+V+ IG L+
Sbjct 682 NPLEPEMMSDEYYIAAAAKRRGLGLVRFIGYLY 714
> At1g62410
Length=223
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 35/103 (33%), Positives = 50/103 (48%), Gaps = 16/103 (15%)
Query 37 IIDKAVTEPDWSEMYADLC-----QLLQWRSVAPEGDPETIRRTPFMLGLLTRIQQEFEA 91
I DKAV EP + MYA LC ++ ++ AP+ D + +R LL Q+ FE
Sbjct 17 IFDKAVLEPTFCPMYAQLCFDIRHKMPRFPPSAPKTDEISFKRV-----LLNTCQKVFER 71
Query 92 MPAALQHEVEG----EAEEALQEEVR-LKRRVLGVVKLIGELF 129
L E+ + E ++EVR L R LG ++ GELF
Sbjct 72 T-DDLSEEIRKMNAPDQEAEREDEVRLLNLRTLGNLRFCGELF 113
> At5g57870
Length=780
Score = 41.6 bits (96), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 34/97 (35%), Positives = 47/97 (48%), Gaps = 15/97 (15%)
Query 1 LNKLTIEKFGVIAEKLAQILEKSLNCQAEIRIQVDAIIDKAVTEPDWSEMYADLC----- 55
LNKLT EK+ ++ Q++E + ++ + I DKAV EP + MYA LC
Sbjct 223 LNKLTPEKYDLLK---GQLIESGITSADILKGVITLIFDKAVLEPTFCPMYAKLCSDIND 279
Query 56 QLLQWRSVAPEGDPE-TIRRTPFMLGLLTRIQQEFEA 91
QL + P GD E T +R LL Q+ FE
Sbjct 280 QLPTFPPAEP-GDKEITFKRV-----LLNICQEAFEG 310
> 7304373
Length=1666
Score = 41.2 bits (95), Expect = 6e-04, Method: Composition-based stats.
Identities = 28/92 (30%), Positives = 50/92 (54%), Gaps = 6/92 (6%)
Query 1 LNKLTIEKFGVIAEKLAQILEKSLNCQAEIRIQVDAIIDKAVTEPDWSEMYADLCQLL-- 58
LNKLT E+F + E +I++ ++ ++ + + +KA+ EP++S YA LCQ L
Sbjct 757 LNKLTPERFDTLVE---EIIKLKIDTPDKVDEVIVLVFEKAIDEPNFSVSYARLCQRLAA 813
Query 59 QWRSVAPEGDPETIRRTP-FMLGLLTRIQQEF 89
+ + + + ET + F LL + +QEF
Sbjct 814 EVKVIDERMESETKSNSAHFRNALLDKTEQEF 845
> YGR162w
Length=952
Score = 40.4 bits (93), Expect = 7e-04, Method: Composition-based stats.
Identities = 44/146 (30%), Positives = 63/146 (43%), Gaps = 22/146 (15%)
Query 1 LNKLTIEKFGVIAEKLAQILEKSL--NCQAEIRIQVDAIIDKAVTEPDWSEMYADLCQLL 58
LNKLT+E F I+ ++ I S+ ++ ++ I KA EP WS MYA LC
Sbjct 614 LNKLTLEMFDAISSEILAIANISVWETNGETLKAVIEQIFLKACDEPHWSSMYAQLCG-- 671
Query 59 QWRSVAPEGDPETIRRTP--------FMLGLLTRIQQEFE-AMPAALQHEVEGEA--EEA 107
V E +P+ T + L+ R EF+ L +G E
Sbjct 672 ---KVVKELNPDITDETNEGKTGPKLVLHYLVARCHAEFDKGWTDKLPTNEDGTPLEPEM 728
Query 108 LQEE----VRLKRRVLGVVKLIGELF 129
+ EE KRR LG+V+ IG L+
Sbjct 729 MSEEYYAAASAKRRGLGLVRFIGFLY 754
> CE02277
Length=1156
Score = 37.7 bits (86), Expect = 0.005, Method: Composition-based stats.
Identities = 29/111 (26%), Positives = 53/111 (47%), Gaps = 14/111 (12%)
Query 28 AEIRIQVDAIIDKAVTEPDWSEMYADLCQLLQWRSVAPEGDPETIRRTPFMLGLLTRIQQ 87
A++ V+ + DKAV EP + +YA++C+ ++ G R +LTR Q
Sbjct 550 AQLAQVVEIVFDKAVEEPKFCALYAEMCKAQANHELSQTGGKSAFRNK-----VLTRTQM 604
Query 88 EFE------AMPAALQHEVEGEAEEAL---QEEVRLKRRVLGVVKLIGELF 129
F+ A A+ + E ++ L +E+ + +RR GV+ +G L+
Sbjct 605 TFQDKKDIDADKLAIIEKEEDPVKKELMLAEEKQKFRRRKFGVMTFMGYLY 655
> 7303385
Length=869
Score = 33.1 bits (74), Expect = 0.15, Method: Composition-based stats.
Identities = 27/82 (32%), Positives = 38/82 (46%), Gaps = 5/82 (6%)
Query 50 MYADLCQLLQWRSVAPEGDPETIRRTPFMLGLLTRIQQEFEAMPAALQHEVEGEAEEALQ 109
MYA LC+ L AP D E + F+ L+ + +F +++ E
Sbjct 1 MYAQLCKRLS--EEAPSFDKEPSNSSTFLRLLIAVCRDKFNNRLKRDENDNRPPPENEAD 58
Query 110 EEVR---LKRRVLGVVKLIGEL 128
EE R K+R+LG VK IGEL
Sbjct 59 EEERRHLAKQRMLGNVKFIGEL 80
> SPAC1006.05c
Length=396
Score = 31.2 bits (69), Expect = 0.46, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 17/33 (51%), Gaps = 3/33 (9%)
Query 45 PDWSEMYADLCQLLQWRSVAPEGDP---ETIRR 74
PDW++ YA Q QW A G P E +RR
Sbjct 264 PDWNDYYARRVQFCQWTIAAAPGHPILWELVRR 296
> 7297353
Length=1216
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 14/47 (29%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 69 PETIRRTPFMLGLLTRIQQEFEAMPAALQHEVEGEAEEALQEEVRLK 115
PE+ TP G+L+R Q E+E + ++ +G +E + E+ L+
Sbjct 1158 PESSFTTPRRRGVLSRTQSEWEEVYPVVKFNRQGSSESDISEDNELR 1204
> CE22949
Length=477
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 29/46 (63%), Gaps = 2/46 (4%)
Query 41 AVTEPDW-SEMYADLCQL-LQWRSVAPEGDPETIRRTPFMLGLLTR 84
V +PDW + + A++ +L Q+RS+ D ++ RT ++LG +T+
Sbjct 188 GVAQPDWVNHVLANVTELKRQYRSIQFNSDEKSKMRTGYLLGQITK 233
> ECU08g1370
Length=244
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 21/79 (26%), Positives = 38/79 (48%), Gaps = 8/79 (10%)
Query 53 DLCQLLQWRSVAPEGDPETIRRTPFMLGLLTRIQQEFEAMPAALQHEV-EGEAEEALQEE 111
D+C+L+++ GD + I+ ++ R+ EFEA P A + G A+ Q +
Sbjct 15 DVCRLVRYF-----GDEQGIKVPEQIVDDYNRVGAEFEASPKAFDYVFHNGNAQNIFQYK 69
Query 112 VRLKRRVLGVVKLIGELFH 130
R ++ V L+ + FH
Sbjct 70 DR--EELIASVFLVRDTFH 86
> At5g48160
Length=574
Score = 28.9 bits (63), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 12/34 (35%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 44 EPDWSEMYADLCQLLQWRSVAPEGDPETIRRTPF 77
+P MY+ + LLQ +++P+ DP+ R PF
Sbjct 537 DPSQVMMYSKIRDLLQGYNLSPKVDPQLNERNPF 570
> YGL038c
Length=480
Score = 28.9 bits (63), Expect = 2.8, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 12/24 (50%), Gaps = 0/24 (0%)
Query 46 DWSEMYADLCQLLQWRSVAPEGDP 69
DWSE YA Q QW A G P
Sbjct 250 DWSEWYARRIQFCQWTIQAKPGHP 273
> Hs4506675
Length=607
Score = 27.7 bits (60), Expect = 5.1, Method: Composition-based stats.
Identities = 20/47 (42%), Positives = 24/47 (51%), Gaps = 4/47 (8%)
Query 67 GDPETIRRTPFMLGLLTRIQQEFEAMPAALQHEVEGEAEEALQEEVR 113
G T R T F+L L + E EA A L +V+GE EE EVR
Sbjct 59 GGGSTSRATSFLLAL----EPELEARLAHLGVQVKGEDEEENNLEVR 101
> ECU04g1610
Length=627
Score = 26.9 bits (58), Expect = 9.9, Method: Composition-based stats.
Identities = 21/77 (27%), Positives = 36/77 (46%), Gaps = 3/77 (3%)
Query 1 LNKLTIEKFGVIAEKLAQILEKSLNCQAEIRIQVDAIIDKAVTEPDWSEMYADLCQLLQW 60
N+LT + G++ + L I ++ EI + DKA++EP + + YA L L+
Sbjct 210 FNRLTAKNIGLVIKNLKAIRVGTIEEMKEI---AKILFDKAISEPTFVKYYALLVLDLKK 266
Query 61 RSVAPEGDPETIRRTPF 77
+ E I +T F
Sbjct 267 EWQSEEEKTRDITQTVF 283
Lambda K H
0.319 0.136 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1246445644
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40