bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1177_orf1
Length=180
Score E
Sequences producing significant alignments: (Bits) Value
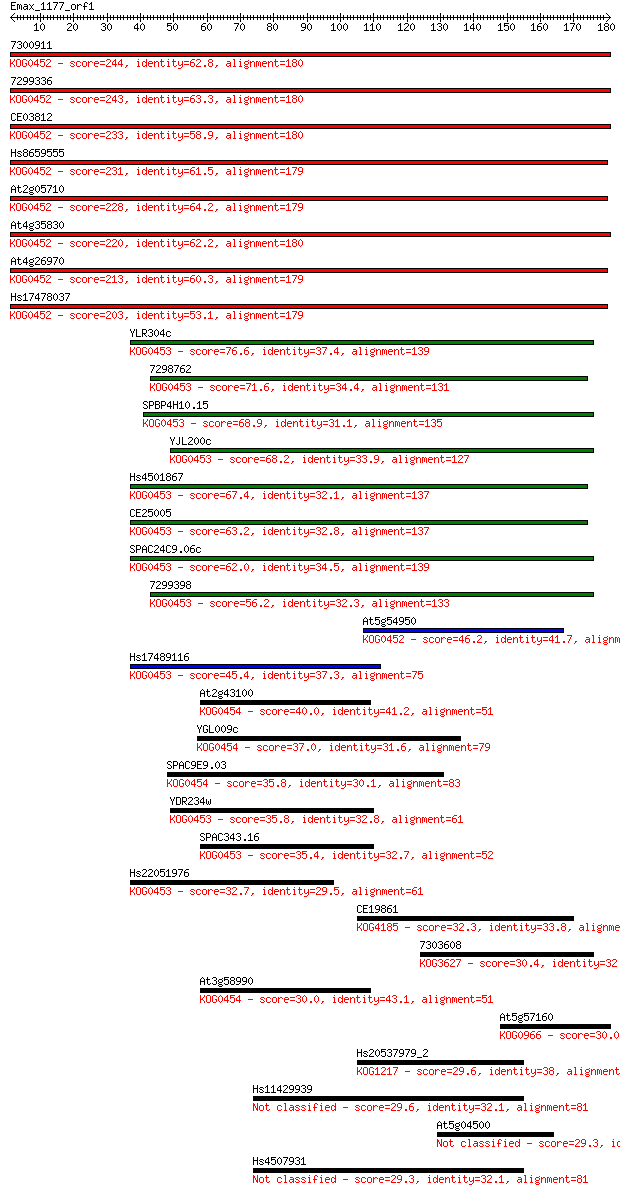
7300911 244 7e-65
7299336 243 2e-64
CE03812 233 2e-61
Hs8659555 231 8e-61
At2g05710 228 5e-60
At4g35830 220 2e-57
At4g26970 213 1e-55
Hs17478037 203 2e-52
YLR304c 76.6 2e-14
7298762 71.6 7e-13
SPBP4H10.15 68.9 5e-12
YJL200c 68.2 8e-12
Hs4501867 67.4 1e-11
CE25005 63.2 3e-10
SPAC24C9.06c 62.0 6e-10
7299398 56.2 4e-08
At5g54950 46.2 3e-05
Hs17489116 45.4 5e-05
At2g43100 40.0 0.002
YGL009c 37.0 0.020
SPAC9E9.03 35.8 0.049
YDR234w 35.8 0.052
SPAC343.16 35.4 0.062
Hs22051976 32.7 0.38
CE19861 32.3 0.53
7303608 30.4 2.1
At3g58990 30.0 2.7
At5g57160 30.0 2.7
Hs20537979_2 29.6 3.4
Hs11429939 29.6 3.8
At5g04500 29.3 3.9
Hs4507931 29.3 4.6
> 7300911
Length=902
Score = 244 bits (623), Expect = 7e-65, Method: Compositional matrix adjust.
Identities = 113/180 (62%), Positives = 142/180 (78%), Gaps = 1/180 (0%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIVL 60
GSRRGND VM RGTFANIRLVNK+ GP +H+P+ E + + + AERY +EG P++++
Sbjct 723 GSRRGNDAVMARGTFANIRLVNKLASKTGPSTLHVPSGEEMDIFDAAERYASEGTPLVLV 782
Query 61 AGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKL 120
GK+YGSGSSRDWAAKGP+L G+KAVIAES+ERIHRSNLVGMGI+PLQFL GQSADTLKL
Sbjct 783 VGKDYGSGSSRDWAAKGPFLLGIKAVIAESYERIHRSNLVGMGIIPLQFLPGQSADTLKL 842
Query 121 TGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLRNML 180
+GRE +NI L G L PG ++V D G FE R DT+V++ Y++NGG L+Y++R ML
Sbjct 843 SGREVYNIVLPEGELKPGQRIQVDAD-GNVFETTLRFDTEVDITYYKNGGILNYMIRKML 901
> 7299336
Length=899
Score = 243 bits (619), Expect = 2e-64, Method: Compositional matrix adjust.
Identities = 114/180 (63%), Positives = 142/180 (78%), Gaps = 1/180 (0%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIVL 60
GSRRGND +M RGTFANIRLVNK+ GP+ +H+P+ E L + + AERYR EG P++++
Sbjct 720 GSRRGNDAIMSRGTFANIRLVNKLVEKTGPRTVHIPSQEELDIFDAAERYREEGTPLVLV 779
Query 61 AGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKL 120
GK+YGSGSSRDWAAKGP+L GVKAVIAES+ERIHRSNLVGMGI+PLQFL GQSA+TL L
Sbjct 780 VGKDYGSGSSRDWAAKGPFLLGVKAVIAESYERIHRSNLVGMGIIPLQFLPGQSAETLNL 839
Query 121 TGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLRNML 180
TGRE +NIAL L PG ++V D G FE R DT+V++ Y++NGG L+Y++R ML
Sbjct 840 TGREVYNIALPESGLKPGQKIQVEAD-GTVFETILRFDTEVDITYYKNGGILNYMIRKML 898
> CE03812
Length=887
Score = 233 bits (593), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 106/180 (58%), Positives = 139/180 (77%), Gaps = 1/180 (0%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIVL 60
G+RRGNDE+M RGTFANIRLVNK+ GP +H+P+ E L + + A++Y+ G P I+L
Sbjct 708 GARRGNDEIMARGTFANIRLVNKLASKVGPITLHVPSGEELDIFDAAQKYKDAGIPAIIL 767
Query 61 AGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKL 120
AGKEYG GSSRDWAAKGP+LQGVKAVIAESFERIHRSNL+GMGI+P Q+ GQ+AD+L L
Sbjct 768 AGKEYGCGSSRDWAAKGPFLQGVKAVIAESFERIHRSNLIGMGIIPFQYQAGQNADSLGL 827
Query 121 TGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLRNML 180
TG+E+F+I + L PG ++ V+ G F+ CR DT+VE+ Y+RNGG L Y++R ++
Sbjct 828 TGKEQFSIGV-PDDLKPGQLIDVNVSNGSVFQVICRFDTEVELTYYRNGGILQYMIRKLI 886
> Hs8659555
Length=889
Score = 231 bits (588), Expect = 8e-61, Method: Compositional matrix adjust.
Identities = 110/179 (61%), Positives = 139/179 (77%), Gaps = 1/179 (0%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIVL 60
GSRRGND VM RGTFANIRL+N+ P+ +HLP+ EIL V + AERY+ G P+IVL
Sbjct 710 GSRRGNDAVMARGTFANIRLLNRFLNKQAPQTIHLPSGEILDVFDAAERYQQAGLPLIVL 769
Query 61 AGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKL 120
AGKEYG+GSSRDWAAKGP+L G+KAV+AES+ERIHRSNLVGMG++PL++L G++AD L L
Sbjct 770 AGKEYGAGSSRDWAAKGPFLLGIKAVLAESYERIHRSNLVGMGVIPLEYLPGENADALGL 829
Query 121 TGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLRNM 179
TG+ER+ I + L P V+V D GK+F+A R DTDVE+ YF NGG L+Y++R M
Sbjct 830 TGQERYTIII-PENLKPQMKVQVKLDTGKTFQAVMRFDTDVELTYFLNGGILNYMIRKM 887
> At2g05710
Length=898
Score = 228 bits (581), Expect = 5e-60, Method: Compositional matrix adjust.
Identities = 115/182 (63%), Positives = 137/182 (75%), Gaps = 3/182 (1%)
Query 1 GSRRGNDEVMVRGTFANIRLVNK-MCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIV 59
GSRRGNDE+M RGTFANIR+VNK M GPK +H+P+ E L V + A RY++ G+ I+
Sbjct 714 GSRRGNDEIMARGTFANIRIVNKLMNGEVGPKTVHIPSGEKLSVFDAAMRYKSSGEDTII 773
Query 60 LAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLK 119
LAG EYGSGSSRDWAAKGP LQGVKAVIA+SFERIHRSNLVGMGI+PL F G+ ADTL
Sbjct 774 LAGAEYGSGSSRDWAAKGPMLQGVKAVIAKSFERIHRSNLVGMGIIPLCFKSGEDADTLG 833
Query 120 LTGRERFNIAL--NGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLR 177
LTG ER+ I L + + PG V V+TD GKSF R DT+VE+ YF +GG L YV+R
Sbjct 834 LTGHERYTIHLPTDISEIRPGQDVTVTTDNGKSFTCTVRFDTEVELAYFNHGGILPYVIR 893
Query 178 NM 179
N+
Sbjct 894 NL 895
> At4g35830
Length=898
Score = 220 bits (560), Expect = 2e-57, Method: Compositional matrix adjust.
Identities = 112/183 (61%), Positives = 135/183 (73%), Gaps = 3/183 (1%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCP-SAGPKAMHLPTNEILPVSEVAERYRAEGQPMIV 59
GSRRGNDE+M RGTFANIR+VNK GPK +H+PT E L V + A +YR EG+ I+
Sbjct 714 GSRRGNDEIMARGTFANIRIVNKHLKGEVGPKTVHIPTGEKLSVFDAAMKYRNEGRDTII 773
Query 60 LAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLK 119
LAG EYGSGSSRDWAAKGP L GVKAVI++SFERIHRSNLVGMGI+PL F G+ A+TL
Sbjct 774 LAGAEYGSGSSRDWAAKGPMLLGVKAVISKSFERIHRSNLVGMGIIPLCFKAGEDAETLG 833
Query 120 LTGRERFNIAL--NGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLR 177
LTG+E + I L N + PG V V T+ GKSF R DT+VE+ YF +GG L YV+R
Sbjct 834 LTGQELYTIELPNNVSEIKPGQDVTVVTNNGKSFTCTLRFDTEVELAYFDHGGILQYVIR 893
Query 178 NML 180
N++
Sbjct 894 NLI 896
> At4g26970
Length=907
Score = 213 bits (543), Expect = 1e-55, Method: Compositional matrix adjust.
Identities = 108/182 (59%), Positives = 131/182 (71%), Gaps = 3/182 (1%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCP-SAGPKAMHLPTNEILPVSEVAERYRAEGQPMIV 59
GSRRGNDEVM RGTFANIR+VNK+ GP +H+PT E L V + A +Y+ Q I+
Sbjct 723 GSRRGNDEVMARGTFANIRIVNKLLKGEVGPNTVHIPTGEKLSVFDAASKYKTAEQDTII 782
Query 60 LAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLK 119
LAG EYGSGSSRDWAAKGP L GVKAVIA+SFERIHRSNL GMGI+PL F G+ A+TL
Sbjct 783 LAGAEYGSGSSRDWAAKGPLLLGVKAVIAKSFERIHRSNLAGMGIIPLCFKAGEDAETLG 842
Query 120 LTGRERFNIALNG--GRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLR 177
LTG ER+ + L + PG V V+TD GKSF R DT+VE+ Y+ +GG L YV+R
Sbjct 843 LTGHERYTVHLPTKVSDIRPGQDVTVTTDSGKSFVCTLRFDTEVELAYYDHGGILPYVIR 902
Query 178 NM 179
++
Sbjct 903 SL 904
> Hs17478037
Length=963
Score = 203 bits (516), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 95/179 (53%), Positives = 127/179 (70%), Gaps = 1/179 (0%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIVL 60
G+RRGND VM RGTFANI+L NK PK +H P+ + L V E AE Y+ EG P+I+L
Sbjct 785 GARRGNDAVMTRGTFANIKLFNKFIGKPAPKTIHFPSGQTLDVFEAAELYQKEGIPLIIL 844
Query 61 AGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKL 120
AGK+YGSG+SRDWAAKGPYL GVKAV+AES+E+IH+ +L+G+GI PLQFL G++AD+L L
Sbjct 845 AGKKYGSGNSRDWAAKGPYLLGVKAVLAESYEKIHKDHLIGIGIAPLQFLPGENADSLGL 904
Query 121 TGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLRNM 179
+GRE F++ L PG + + T GK F + DVE+ +++GG L++V R
Sbjct 905 SGRETFSLTFP-EELSPGITLNIQTSTGKVFSVIASFEDDVEITLYKHGGLLNFVARKF 962
> YLR304c
Length=778
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 52/142 (36%), Positives = 74/142 (52%), Gaps = 6/142 (4%)
Query 37 TNEILPVSEVAERYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHR 96
T E V + A YR +G +V+ + +G GSSR+ AA P G A+I +SF RIH
Sbjct 634 TGEYKGVPDTARDYRDQGIKWVVIGDENFGEGSSREHAALEPRFLGGFAIITKSFARIHE 693
Query 97 SNLVGMGILPLQFLEGQSADTLKLTGRERFNIALNGGRLVPGSIV--RVSTDCGKSFEAK 154
+NL G+LPL F AD K+ +R +I L L PG V RV GK ++A
Sbjct 694 TNLKKQGLLPLNF--KNPADYDKINPDDRIDI-LGLAELAPGKPVTMRVHPKNGKPWDAV 750
Query 155 -CRIDTDVEVEYFRNGGALHYV 175
D ++E+F+ G AL+ +
Sbjct 751 LTHTFNDEQIEWFKYGSALNKI 772
> 7298762
Length=683
Score = 71.6 bits (174), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 45/134 (33%), Positives = 69/134 (51%), Gaps = 7/134 (5%)
Query 43 VSEVAERYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGM 102
V +VA Y+A G + + + YG GSSR+ AA P G +A+I +SF RIH +NL
Sbjct 547 VPDVARDYKANGIKWVAVGDENYGEGSSREHAALEPRHLGGRAIIVKSFARIHETNLKKQ 606
Query 103 GILPLQFLEGQSADTLKLTGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRID---T 159
G+LPL F D ++ T + LN L PG V G E + +++
Sbjct 607 GLLPLTFANPADYDKIQPTSKISL---LNLKSLAPGKPVDAEIKNGDKVE-RIKLNHTLN 662
Query 160 DVEVEYFRNGGALH 173
D+++ +F+ G AL+
Sbjct 663 DLQIGWFKAGSALN 676
> SPBP4H10.15
Length=905
Score = 68.9 bits (167), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 42/141 (29%), Positives = 74/141 (52%), Gaps = 9/141 (6%)
Query 41 LPVSEVAERYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLV 100
+ + E+ +++ +GQP +V+A YG GS+R+ AA P + ++ +SF RIH +NL
Sbjct 645 MTIPELMWKWKKDGQPWLVVAEHNYGEGSAREHAALQPRAMNGRIILTKSFARIHETNLK 704
Query 101 GMGILPLQFLEGQSADTLKLTGRERFNI-----ALNGGRLVPGSIVRVSTDCGKSFEAKC 155
G+LPL F+ AD K+ ++ + L G P ++V D G E C
Sbjct 705 KQGVLPLTFV--NEADYEKIDAEDKVSTRGIEQLLEGVLDQPITLVVTKKD-GSVVEIPC 761
Query 156 R-IDTDVEVEYFRNGGALHYV 175
+ + ++E+F+ G AL+ +
Sbjct 762 KHTMSKDQIEFFKAGSALNLI 782
> YJL200c
Length=789
Score = 68.2 bits (165), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 43/138 (31%), Positives = 76/138 (55%), Gaps = 17/138 (12%)
Query 49 RYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQ 108
+++++G+P V+A YG GS+R+ AA P G + ++ +SF RIH +NL G+LPL
Sbjct 651 KWKSDGRPWTVIAEHNYGEGSAREHAALSPRFLGGEILLVKSFARIHETNLKKQGVLPLT 710
Query 109 FLEGQSADTLKLTGR--ERFNIAL-------NGGRLVPGSIVRVSTDCGKSF--EAKCRI 157
F D + +G E N+ NGG + V+++ G+SF +AK +
Sbjct 711 FANESDYDKIS-SGDVLETLNLVDMIAKDGNNGGEID----VKITKPNGESFTIKAKHTM 765
Query 158 DTDVEVEYFRNGGALHYV 175
D ++++F+ G A++Y+
Sbjct 766 SKD-QIDFFKAGSAINYI 782
> Hs4501867
Length=780
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 44/140 (31%), Positives = 69/140 (49%), Gaps = 6/140 (4%)
Query 37 TNEILPVSEVAERYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHR 96
T E PV + A Y+ G +V+ + YG GSSR+ AA P G +A+I +SF RIH
Sbjct 637 TQEFGPVPDTARYYKKHGIRWVVIGDENYGEGSSREHAALEPRHLGGRAIITKSFARIHE 696
Query 97 SNLVGMGILPLQFLEGQSADTLKLTGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCR 156
+NL G+LPL F + AD K+ ++ I PG ++ +
Sbjct 697 TNLKKQGLLPLTFAD--PADYNKIHPVDKLTIQ-GLKDFTPGKPLKCIIKHPNGTQETIL 753
Query 157 ID---TDVEVEYFRNGGALH 173
++ + ++E+FR G AL+
Sbjct 754 LNHTFNETQIEWFRAGSALN 773
> CE25005
Length=777
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 45/141 (31%), Positives = 71/141 (50%), Gaps = 8/141 (5%)
Query 37 TNEILPVSEVAERYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHR 96
T E V A +Y+A+G + + + YG GSSR+ AA P G +A+I +SF RIH
Sbjct 631 TGEYGAVPATARKYKADGVRWVAIGDENYGEGSSREHAALEPRHLGGRAIIVKSFARIHE 690
Query 97 SNLVGMGILPLQFLEGQSADTLKLTGRERFNIALNG-GRLVPG---SIVRVSTDCGKSFE 152
+NL G+LPL F AD K+ + N+++ G PG + + T+ K
Sbjct 691 TNLKKQGMLPLTF--ANPADYDKIDPSD--NVSIVGLSSFAPGKPLTAIFKKTNGSKVEV 746
Query 153 AKCRIDTDVEVEYFRNGGALH 173
+ ++E+F+ G AL+
Sbjct 747 TLNHTFNEQQIEWFKAGSALN 767
> SPAC24C9.06c
Length=778
Score = 62.0 bits (149), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 48/147 (32%), Positives = 71/147 (48%), Gaps = 15/147 (10%)
Query 37 TNEILPVSEVAERYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHR 96
T E V VA YR G + L + +G GSSR+ AA P G AVI +SF RIH
Sbjct 632 TGEYKTVPNVAIDYRDHGIRWVTLGEQNFGEGSSREHAALEPRYLGGAAVITKSFARIHE 691
Query 97 SNLVGMGILPLQFLEGQSA------DTLKLTGRERFNIALNGGRLVPGSIVRVSTDCGKS 150
+NL G+LPL F + + DT+ + G F G+ P ++V D
Sbjct 692 TNLKKQGLLPLTFADPAAYDKISPFDTVDIDGLTTF----APGK--PLTLVVHPADGSAE 745
Query 151 FEAKCR--IDTDVEVEYFRNGGALHYV 175
+ K + D ++E+F+ G AL+++
Sbjct 746 WSTKLNHTFNKD-QIEWFKAGSALNHM 771
> 7299398
Length=769
Score = 56.2 bits (134), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 43/136 (31%), Positives = 67/136 (49%), Gaps = 7/136 (5%)
Query 43 VSEVAERYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGM 102
V VA Y+A + + YG GSSR+ AA P G A+I +SF RIH +NL
Sbjct 627 VPAVARDYKANKIKWCAVGEENYGEGSSREHAALEPRHLGGVAIIVKSFARIHETNLKKQ 686
Query 103 GILPLQFLEGQSADTLKLTGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRID---T 159
G+L L F D K+ + +I LN L PG V S + K +++
Sbjct 687 GMLALTF--ANPGDYDKVQPSSKISI-LNLKDLAPGKPVDAEIKNNGSSD-KIQLNHTLN 742
Query 160 DVEVEYFRNGGALHYV 175
++++++F+ G AL+ +
Sbjct 743 ELQIQWFQAGSALNLM 758
> At5g54950
Length=74
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 35/62 (56%), Gaps = 2/62 (3%)
Query 107 LQFLEGQSADTLKLTGRERFNIAL--NGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVE 164
+ F G+ A+TL LTG E + I L N + PG + V+TD KSF R+DT++ V
Sbjct 1 MAFKSGEDAETLGLTGHELYTIHLPSNINEIKPGQDITVTTDTAKSFVCTLRLDTEIWVL 60
Query 165 YF 166
F
Sbjct 61 CF 62
> Hs17489116
Length=716
Score = 45.4 bits (106), Expect = 5e-05, Method: Composition-based stats.
Identities = 28/76 (36%), Positives = 43/76 (56%), Gaps = 6/76 (7%)
Query 37 TNEILPVSEVAERYRAEGQPMIVLAGKEYGSGSSRDWAA-KGPYLQGVKAVIAESFERIH 95
T E PV + A Y+ G +V+ + SSR+ A + P+L G +A+I +SF RIH
Sbjct 577 TQEFGPVPDTARYYKKHGIRWVVIGDEN----SSREHAVLEPPHLWG-QAIITKSFARIH 631
Query 96 RSNLVGMGILPLQFLE 111
++N G+LPL F +
Sbjct 632 KTNPKKQGLLPLTFAD 647
> At2g43100
Length=256
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 31/52 (59%), Gaps = 1/52 (1%)
Query 58 IVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMG-ILPLQ 108
I++ G+ +G GSSR+ A G KA++AES+ RI N V G + PL+
Sbjct 139 IIIGGENFGCGSSREHAPVCLGAAGAKAIVAESYARIFFRNSVATGEVFPLE 190
> YGL009c
Length=779
Score = 37.0 bits (84), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 40/79 (50%), Gaps = 7/79 (8%)
Query 57 MIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSAD 116
++V+ G +G GSSR+ A G+K++IA S+ I +N G+LP++ + D
Sbjct 617 ILVVTGDNFGCGSSREHAPWALKDFGIKSIIAPSYGDIFYNNSFKNGLLPIRLDQQIIID 676
Query 117 TLKLTGRERFNIALNGGRL 135
L IA GG+L
Sbjct 677 KL-------IPIANKGGKL 688
> SPAC9E9.03
Length=758
Score = 35.8 bits (81), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 25/84 (29%), Positives = 41/84 (48%), Gaps = 4/84 (4%)
Query 48 ERYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPL 107
E YR +++A +G GSSR+ A G++ +IA SF I +N G+LP+
Sbjct 599 EPYR---HATVLVAHDNFGCGSSREHAPWALNDFGIRVIIAPSFADIFFNNCFKNGMLPI 655
Query 108 QFLEGQSADTLKLTGRE-RFNIAL 130
Q D +K + +F++ L
Sbjct 656 PTPIEQVNDMMKAAENQVKFSVDL 679
> YDR234w
Length=693
Score = 35.8 bits (81), Expect = 0.052, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 34/62 (54%), Gaps = 1/62 (1%)
Query 49 RYRAEGQP-MIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPL 107
+R + P IV++G +G+GSSR+ AA +G+ V++ SF I N + +L L
Sbjct 556 EFRTKVHPGDIVVSGFNFGTGSSREQAATALLAKGINLVVSGSFGNIFSRNSINNALLTL 615
Query 108 QF 109
+
Sbjct 616 EI 617
> SPAC343.16
Length=721
Score = 35.4 bits (80), Expect = 0.062, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 30/52 (57%), Gaps = 0/52 (0%)
Query 58 IVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQF 109
I+++G +G+GSSR+ AA +G+ V+ SF I + N + +L +Q
Sbjct 593 ILVSGFNFGTGSSREQAATAILSRGIPLVVGGSFSDIFKRNSINNALLAIQL 644
> Hs22051976
Length=293
Score = 32.7 bits (73), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 18/61 (29%), Positives = 30/61 (49%), Gaps = 0/61 (0%)
Query 37 TNEILPVSEVAERYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHR 96
T E P+ + A Y+ G +V+ + Y S + A +L +A+I +SF RIH
Sbjct 227 TQEFGPIPDTARYYKKHGIRWVVIRDENYREDSHGEHTALEHHLLRGRAIITKSFARIHE 286
Query 97 S 97
+
Sbjct 287 T 287
> CE19861
Length=261
Score = 32.3 bits (72), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 31/67 (46%), Gaps = 2/67 (2%)
Query 105 LPLQFLEGQSADTLKLTGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRIDTD--VE 162
L L+ L G+S DTL T + + R+ P V DC + F C+ + D VE
Sbjct 195 LILEVLNGKSKDTLSCTSCQLTYSSQTFPRIHPSCGHSVCEDCKEPFCKICKTEEDRVVE 254
Query 163 VEYFRNG 169
+ RNG
Sbjct 255 MILVRNG 261
> 7303608
Length=256
Score = 30.4 bits (67), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 27/52 (51%), Gaps = 1/52 (1%)
Query 124 ERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYV 175
+R+ GG + IV + C +S EAK + V Y+R+GG++H V
Sbjct 49 QRYGSHFCGGSIYSHDIVITAAHCLQSIEAK-DLKIRVGSTYWRSGGSVHSV 99
> At3g58990
Length=253
Score = 30.0 bits (66), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 22/52 (42%), Positives = 30/52 (57%), Gaps = 1/52 (1%)
Query 58 IVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMG-ILPLQ 108
+++ G +G GSSR+ A G KAV+AES+ RI N V G I PL+
Sbjct 136 VIIGGDNFGCGSSREHAPVCLGAAGAKAVVAESYARIFFRNCVATGEIFPLE 187
> At5g57160
Length=1184
Score = 30.0 bits (66), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 19/33 (57%), Gaps = 1/33 (3%)
Query 148 GKSFEAKCRIDTDVEVEYFRNGGALHYVLRNML 180
GK A+C+ D D ++ +NG +HY RN L
Sbjct 245 GKDVVAECKFDGD-RIQIHKNGTDIHYFSRNFL 276
> Hs20537979_2
Length=1158
Score = 29.6 bits (65), Expect = 3.4, Method: Composition-based stats.
Identities = 19/50 (38%), Positives = 23/50 (46%), Gaps = 1/50 (2%)
Query 105 LPLQFLEGQSADTLKLTGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAK 154
L LQ E S D + G N NGG VPG+ S DCG F+ +
Sbjct 1037 LALQLPEHGSKDIGNVPGNCSENPCQNGGTCVPGADAH-SCDCGPGFKGR 1085
> Hs11429939
Length=351
Score = 29.6 bits (65), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 26/87 (29%), Positives = 35/87 (40%), Gaps = 6/87 (6%)
Query 74 AAKGPYLQGV------KAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKLTGRERFN 127
A K LQG + IA++F I LV + P LE ++ L GRE
Sbjct 215 ALKVDLLQGAGNSAAGRGAIADTFRSISTRELVHLEDSPDYCLENKTLGLLGTEGRECLR 274
Query 128 IALNGGRLVPGSIVRVSTDCGKSFEAK 154
GR S R+ DCG + E +
Sbjct 275 RGRALGRWERRSCRRLCGDCGLAVEER 301
> At5g04500
Length=764
Score = 29.3 bits (64), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 24/37 (64%), Gaps = 3/37 (8%)
Query 129 ALNGGR--LVPGSIVRVSTDCGKSFEAKCRIDTDVEV 163
A NGGR L GS+ RV DCG+++ + R+ + +EV
Sbjct 297 ARNGGRAFLYDGSLYRVGQDCGENYGKRIRV-SKIEV 332
> Hs4507931
Length=351
Score = 29.3 bits (64), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 26/87 (29%), Positives = 35/87 (40%), Gaps = 6/87 (6%)
Query 74 AAKGPYLQGV------KAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKLTGRERFN 127
A K LQG + IA++F I LV + P LE ++ L GRE
Sbjct 215 ALKVDLLQGAGNSAAARGAIADTFRSISTRELVHLEDSPDYCLENKTLGLLGTEGRECLR 274
Query 128 IALNGGRLVPGSIVRVSTDCGKSFEAK 154
GR S R+ DCG + E +
Sbjct 275 RGRALGRWELRSCRRLCGDCGLAVEER 301
Lambda K H
0.319 0.137 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2806646388
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40