bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1143_orf2
Length=387
Score E
Sequences producing significant alignments: (Bits) Value
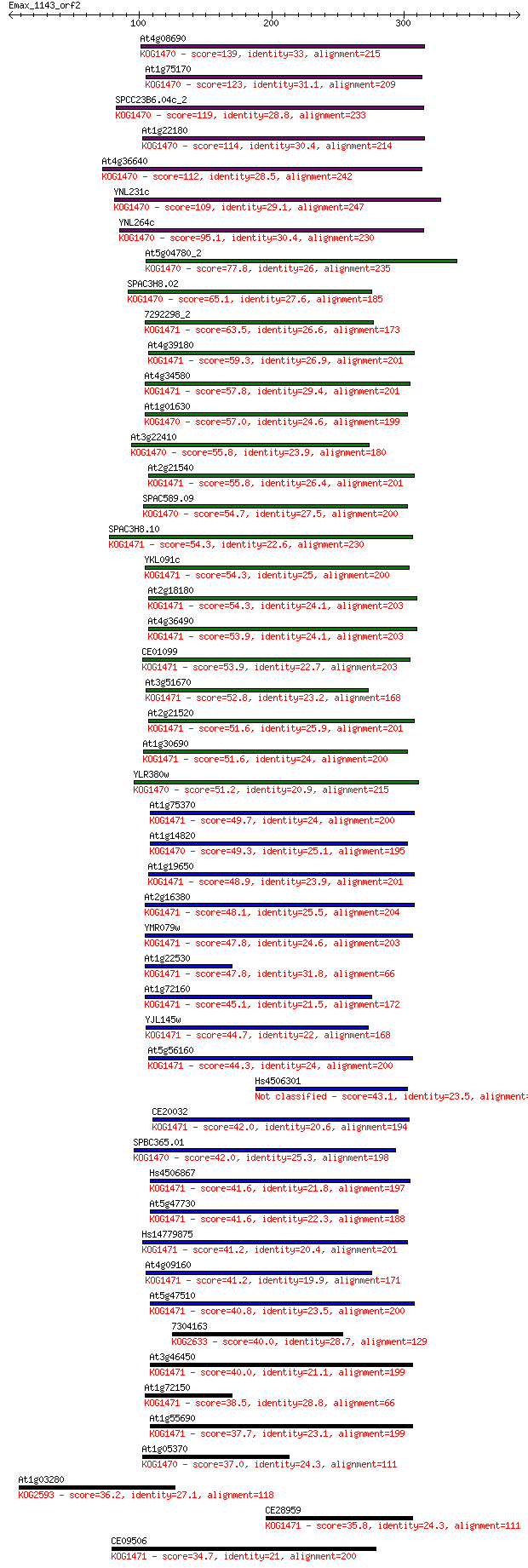
At4g08690 139 9e-33
At1g75170 123 7e-28
SPCC23B6.04c_2 119 9e-27
At1g22180 114 2e-25
At4g36640 112 1e-24
YNL231c 109 1e-23
YNL264c 95.1 3e-19
At5g04780_2 77.8 4e-14
SPAC3H8.02 65.1 3e-10
7292298_2 63.5 8e-10
At4g39180 59.3 2e-08
At4g34580 57.8 4e-08
At1g01630 57.0 7e-08
At3g22410 55.8 1e-07
At2g21540 55.8 2e-07
SPAC589.09 54.7 3e-07
SPAC3H8.10 54.3 4e-07
YKL091c 54.3 4e-07
At2g18180 54.3 5e-07
At4g36490 53.9 5e-07
CE01099 53.9 6e-07
At3g51670 52.8 1e-06
At2g21520 51.6 3e-06
At1g30690 51.6 3e-06
YLR380w 51.2 4e-06
At1g75370 49.7 1e-05
At1g14820 49.3 1e-05
At1g19650 48.9 2e-05
At2g16380 48.1 4e-05
YMR079w 47.8 4e-05
At1g22530 47.8 4e-05
At1g72160 45.1 2e-04
YJL145w 44.7 3e-04
At5g56160 44.3 5e-04
Hs4506301 43.1 0.001
CE20032 42.0 0.002
SPBC365.01 42.0 0.002
Hs4506867 41.6 0.003
At5g47730 41.6 0.003
Hs14779875 41.2 0.004
At4g09160 41.2 0.004
At5g47510 40.8 0.005
7304163 40.0 0.008
At3g46450 40.0 0.009
At1g72150 38.5 0.023
At1g55690 37.7 0.045
At1g05370 37.0 0.082
At1g03280 36.2 0.13
CE28959 35.8 0.16
CE09506 34.7 0.34
> At4g08690
Length=301
Score = 139 bits (350), Expect = 9e-33, Method: Compositional matrix adjust.
Identities = 71/216 (32%), Positives = 123/216 (56%), Gaps = 5/216 (2%)
Query 101 WLSNDLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTAARGSVYRRG- 159
+ S+D +L RYLR+ NW A + L +T++WR + KP I EEV G A G +YR
Sbjct 40 FCSDDAVL-RYLRARNWHVKKATKMLKETLKWRVQYKPEEICWEEVAGEAETGKIYRSSC 98
Query 160 FDVHGRPIVYFKPGREPAQSTRAAQDYTLYTLEKALQSVPVGSGRDQVVFLVDFAGFRVS 219
D GRP++ +P E ++S + Y +Y +E A+Q++P G +Q+V+++DF G+ ++
Sbjct 99 VDKLGRPVLIMRPSVENSKSVKGQIRYLVYCMENAVQNLP--PGEEQMVWMIDFHGYSLA 156
Query 220 QVPSMDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLHPLTAAKVEFINTS 279
V S+ ++E +L EHY + LA A L + P + + W++ R L P T KV+F+ +
Sbjct 157 NV-SLRTTKETAHVLQEHYPERLAFAVLYNPPKFFEPFWKVARPFLEPKTRNKVKFVYSD 215
Query 280 NPKQRSKLTQQIPLHFLEQTLGGTCEAAYNHEVYWE 315
+P + + + + +E GG ++ +N E + E
Sbjct 216 DPNTKVIMEENFDMEKMELAFGGNDDSGFNIEKHSE 251
> At1g75170
Length=296
Score = 123 bits (308), Expect = 7e-28, Method: Compositional matrix adjust.
Identities = 65/210 (30%), Positives = 108/210 (51%), Gaps = 3/210 (1%)
Query 105 DLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTAARGSVYRRGF-DVH 163
D L RYL + NW A++ L +T++WR KP I+ EV G G VY+ GF D H
Sbjct 44 DACLKRYLEARNWNVGKAKKMLEETLKWRSSFKPEEIRWNEVSGEGETGKVYKAGFHDRH 103
Query 164 GRPIVYFKPGREPAQSTRAAQDYTLYTLEKALQSVPVGSGRDQVVFLVDFAGFRVSQVPS 223
GR ++ +PG + +S + +Y +E A+ ++P ++Q+ +L+DF G+ +S
Sbjct 104 GRTVLILRPGLQNTKSLENQMKHLVYLIENAILNLP--EDQEQMSWLIDFTGWSMSTSVP 161
Query 224 MDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLHPLTAAKVEFINTSNPKQ 283
+ +RE + IL HY + LA AFL + P + W++V+ + T KV+F+ N +
Sbjct 162 IKSARETINILQNHYPERLAVAFLYNPPRLFEAFWKIVKYFIDAKTFVKVKFVYPKNSES 221
Query 284 RSKLTQQIPLHFLEQTLGGTCEAAYNHEVY 313
++ L GG YN+E +
Sbjct 222 VELMSTFFDEENLPTEFGGKALLQYNYEEF 251
> SPCC23B6.04c_2
Length=442
Score = 119 bits (299), Expect = 9e-27, Method: Compositional matrix adjust.
Identities = 67/233 (28%), Positives = 122/233 (52%), Gaps = 14/233 (6%)
Query 82 SSGSGSKKEVRELTWEEVLWLSNDLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHI 141
+S S K ++ EL E LWL+ + IL RYLR+ W + A+++++ T+ WRR +++
Sbjct 50 ASNSSKKTDLIEL---ERLWLTRECIL-RYLRATKWHVSNAKKRIVDTLVWRRHFGVNNM 105
Query 142 QPEEVMGTAARGSVYRRGFDVHGRPIVYFKPGREPAQSTRAAQDYTLYTLEKALQSVPVG 201
P+E+ A G G+D GRP +Y P R+ +++ + +++LE A+ +P
Sbjct 106 DPDEIQEENATGKQVLLGYDKDGRPCLYLYPARQNTKTSPLQIRHLVFSLECAIDLMP-- 163
Query 202 SGRDQVVFLVDFAGFRVSQVPSMDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLV 261
G + + L++F PS+ +EV+ IL HY + L +A +++ P + G ++L+
Sbjct 164 PGVETLALLINFKSSSNRSNPSVGQGKEVLNILQTHYCERLGRALVINIPWAVWGFFKLI 223
Query 262 RLMLHPLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGTCEAAYNHEVYW 314
+ P+T K++F L + +P L+ GG+ Y+HE YW
Sbjct 224 SPFIDPITREKLKF--------NEPLDRYVPKDQLDSNFGGSLHFEYHHEKYW 268
> At1g22180
Length=314
Score = 114 bits (286), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 65/216 (30%), Positives = 113/216 (52%), Gaps = 5/216 (2%)
Query 102 LSNDLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTAARGSVYRRGF- 160
+D + RYL + N A + L +T++WR + KP I+ EE+ A G +YR
Sbjct 43 FCSDAAITRYLAARNGHVKKATKMLKETLKWRAQYKPEEIRWEEIAREAETGKIYRANCT 102
Query 161 DVHGRPIVYFKPGREPAQSTRAAQDYTLYTLEKALQSVPVGSGRDQVVFLVDFAGFRVSQ 220
D +GR ++ +P + +S + +Y +E A+ ++P ++Q+V+L+DF GF +S
Sbjct 103 DKYGRTVLVMRPSCQNTKSYKGQIRILVYCMENAILNLP--DNQEQMVWLIDFHGFNMSH 160
Query 221 VPSMDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLHPLTAAKVEFINTSN 280
+ S+ VSRE +L EHY + L A + + P + +++V+ L P T+ KV+F+ + +
Sbjct 161 I-SLKVSRETAHVLQEHYPERLGLAIVYNPPKIFESFYKMVKPFLEPKTSNKVKFVYSDD 219
Query 281 PKQRSKLTQQIPLHFLEQTLGG-TCEAAYNHEVYWE 315
L + LE GG +A +N E Y E
Sbjct 220 NLSNKLLEDLFDMEQLEVAFGGKNSDAGFNFEKYAE 255
> At4g36640
Length=294
Score = 112 bits (280), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 69/250 (27%), Positives = 119/250 (47%), Gaps = 10/250 (4%)
Query 72 IFSRSNSSS-SSSGSGSKKEVRELTWE------EVLWLSNDLILWRYLRSYNWEEAAAQQ 124
+F R N+ + S +VREL L +D L R+L + NW+ A++
Sbjct 1 MFRRRNAHQLDNDDSQQDNKVRELKSAIGPLSGHSLVFCSDASLRRFLDARNWDVEKAKK 60
Query 125 QLMQTVQWRRERKPHHIQPEEVMGTAARGSVYRRGF-DVHGRPIVYFKPGREPAQSTRAA 183
+ +T++WR KP I+ +V G R F D GR ++ +P + + S
Sbjct 61 MIQETLKWRSTYKPQEIRWNQVAHEGETGKASRASFHDRQGRVVLIMRPAMQNSTSQEGN 120
Query 184 QDYTLYTLEKALQSVPVGSGRDQVVFLVDFAGFRVSQVPSMDVSREVVQILNEHYTDILA 243
+ +Y LE A+ ++P G+ Q+ +L+DF G+ ++ P M +RE++ IL +Y + L
Sbjct 121 IRHLVYLLENAIINLP--KGQKQMSWLIDFTGWSMAVNPPMKTTREIIHILQNYYPERLG 178
Query 244 KAFLLDAPGYLDGIWRLVRLMLHPLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGT 303
AFL + P ++R + L P TA KV+F+ + +T + L + GG
Sbjct 179 IAFLYNPPRLFQAVYRAAKYFLDPRTAEKVKFVYPKDKASDELMTTHFDVENLPKEFGGE 238
Query 304 CEAAYNHEVY 313
Y+HE +
Sbjct 239 ATLEYDHEDF 248
> YNL231c
Length=351
Score = 109 bits (273), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 72/262 (27%), Positives = 124/262 (47%), Gaps = 26/262 (9%)
Query 81 SSSGSGSKKEVRELTWEEVLWLSNDLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHH 140
+S + S+ +++ L EE WL+ + L RYLR+ W ++ T+ WRRE H
Sbjct 64 TSEKNKSEDDLKPLEEEEKAWLTRECFL-RYLRATKWVLKDCIDRITMTLAWRREFGISH 122
Query 141 IQPEE--------VMGTAARGSVYRRGFDVHGRPIVYFKPGREPAQSTRAAQDYTLYTLE 192
+ E V G G++ RPI+Y KPGR+ +++ + ++ LE
Sbjct 123 LGEEHGDKITADLVAVENESGKQVILGYENDARPILYLKPGRQNTKTSHRQVQHLVFMLE 182
Query 193 KALQSVPVGSGRDQVVFLVDFAGF----RV---SQVPSMDVSREVVQILNEHYTDILAKA 245
+ + +P +G+D + L+DF + +V S++P + V +EV+ IL HY + L KA
Sbjct 183 RVIDFMP--AGQDSLALLIDFKDYPDVPKVPGNSKIPPIGVGKEVLHILQTHYPERLGKA 240
Query 246 FLLDAPGYLDGIWRLVRLMLHPLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGTCE 305
L + P +L+ + PLT K+ F + +P + L+ GG +
Sbjct 241 LLTNIPWLAWTFLKLIHPFIDPLTREKLVF--------DEPFVKYVPKNELDSLYGGDLK 292
Query 306 AAYNHEVYWEAEAAYHQQIQSH 327
YNH+VYW A ++ + H
Sbjct 293 FKYNHDVYWPALVETAREKRDH 314
> YNL264c
Length=350
Score = 95.1 bits (235), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 70/246 (28%), Positives = 111/246 (45%), Gaps = 31/246 (12%)
Query 85 SGSKKEVRELTWEEVLWLSNDLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPE 144
+G+ R L+ E WLS + L RYLR+ W A A + L +T+ WRRE H + +
Sbjct 73 NGTHANDRPLSDWEKFWLSRECFL-RYLRANKWNTANAIKGLTKTLVWRREIGLTHGKED 131
Query 145 EVMGTAARGSVYRR-------GFDVHGRPIVYFKPGREPAQSTRAAQDYTLYTLEKALQS 197
+ TA + +V GFD RP+ Y K GR+ +S+ +Y +E A
Sbjct 132 KDPLTADKVAVENETGKQVILGFDNAKRPLYYMKNGRQNTESSFRQVQELVYMMETATTV 191
Query 198 VPVGSGRDQVVFLVDFAGFR-----VSQVPSMDVSREVVQILNEHYTDILAKAFLLDAPG 252
P G +++ LVDF ++ + P + ++R + ++ +HY + LAK L++ P
Sbjct 192 AP--QGVEKITVLVDFKSYKEPGIITDKAPPISIARMCLNVMQDHYPERLAKCVLINIPW 249
Query 253 YLDGIWRLVRLMLHPLTAAKV----EFINTSNPKQRSKLTQQIPLHFLEQTLGGTCEAAY 308
+ +++ L P T AK F N P Q L+ G + Y
Sbjct 250 FAWAFLKMMYPFLDPATKAKAIFDEPFENHIEPSQ------------LDALYNGLLDFKY 297
Query 309 NHEVYW 314
HEVYW
Sbjct 298 KHEVYW 303
> At5g04780_2
Length=251
Score = 77.8 bits (190), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 61/236 (25%), Positives = 101/236 (42%), Gaps = 40/236 (16%)
Query 105 DLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTAARGSVYRRGF-DVH 163
D L RYL + NW A++ +T++WR G A G VY+ GF D H
Sbjct 34 DACLKRYLEARNWNVGKAKKMFEETLKWRSSE-----------GEA--GKVYKAGFHDRH 80
Query 164 GRPIVYFKPGREPAQSTRAAQDYTLYTLEKALQSVPVGSGRDQVVFLVDFAGFRVSQVPS 223
GR +LY + A+ ++P ++Q+ +L+DF G+ +S
Sbjct 81 GR-----------------TNTKSLYLIANAILNLP--EDQEQMSWLIDFTGWSMSTSVP 121
Query 224 MDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLHPLTAAKVEFINTSNPKQ 283
+ +RE + IL HY + LA AFL + P + W+ H T KV+F+ N +
Sbjct 122 IKSARETINILQNHYPERLAVAFLYNPPRLFEAFWK-----EHAKTFVKVKFVYPKNQES 176
Query 284 RSKLTQQIPLHFLEQTLGGTCEAAYNHEVYWEAEAAYHQQIQSHNTQTLLNMKSNK 339
++ L GG YN+E + ++ +++ N L N +N+
Sbjct 177 VELMSTFFDEENLPTEFGGKALLQYNYEEF--SKQMNQDDVKTANFWGLCNSNNNQ 230
> SPAC3H8.02
Length=444
Score = 65.1 bits (157), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 51/200 (25%), Positives = 89/200 (44%), Gaps = 22/200 (11%)
Query 91 VRELTWEEVLWLSNDLILWRYLRSYNWEEAAAQQQLMQTVQWR-RERK--------PHHI 141
+R W+ V D +L R+LR+ W AA + M+TV WR RE H
Sbjct 110 LRVCFWDAVNCDDPDGLLLRFLRARKWNVEAALEMFMKTVHWRSREMNVGEIVCNADHLD 169
Query 142 QPEEVMGTAARGSVYRRGFDVHGRPIVYFKPGREPAQ--STRAAQDYTLYTLEKA--LQS 197
+ ++ + G + G D H RP+ Y + S + + T++ +E A +
Sbjct 170 KDDDFVRQLRIGKCFIFGEDKHNRPVCYIRARLHKVGDVSPESVERLTVWVMETARLILK 229
Query 198 VPVGSGRDQVVFLVDFAGFRVSQVPSMDVS--REVVQILNEHYTDILAKAFLLDAPGYLD 255
P+ + VVF D F +S +MD + +++ HY + L + + AP
Sbjct 230 PPIETA--TVVF--DMTDFSMS---NMDYGPLKFMIKCFEAHYPECLGECIVHKAPWLFQ 282
Query 256 GIWRLVRLMLHPLTAAKVEF 275
G+W +++ L P+ +KV+F
Sbjct 283 GVWSIIKSWLDPVVVSKVKF 302
> 7292298_2
Length=212
Score = 63.5 bits (153), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 46/181 (25%), Positives = 87/181 (48%), Gaps = 19/181 (10%)
Query 104 NDLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTAARGSVYRRGFDVH 163
ND L RYLR++ + A Q +++T +WR + + + + R D
Sbjct 24 NDFSLRRYLRAFKTTDDAFQA-ILKTNKWRETYGVDKLSEMDRSQLDKKARLLRHR-DCI 81
Query 164 GRPIVYFKPGREPAQSTRAAQD------YTLYTLEKALQSVPVGSGRDQVVFLVDFAGFR 217
GRP++Y PA++ + +D + +Y LE+A + D++ + D A F
Sbjct 82 GRPVIYI-----PAKNHSSERDIDELTRFIVYNLEEACKKC-FEEVTDRLCIVFDLAEFS 135
Query 218 VSQVPSMD--VSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLHPLTAAKVEF 275
S MD + + ++ +L +H+ + L ++++PG IW +R++L TA KV+F
Sbjct 136 TS---CMDYQLVQNLIWLLGKHFPERLGVCLIINSPGLFSTIWPAIRVLLDDNTAKKVKF 192
Query 276 I 276
+
Sbjct 193 V 193
> At4g39180
Length=550
Score = 59.3 bits (142), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 54/222 (24%), Positives = 96/222 (43%), Gaps = 27/222 (12%)
Query 107 ILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHI-------QPEEVMGTAARGSVYRRG 159
++ R+LR+ ++ A+Q + WR+E I + EEV+ +G G
Sbjct 96 MMLRFLRARKFDLEKAKQMWSDMLNWRKEYGADTIMEDFDFKEIEEVVKYYPQGY---HG 152
Query 160 FDVHGRPIVYFKPGREPAQSTRAAQ------DYTLYTLEKALQ----SVPVGSGR--DQV 207
D GRPI + G+ A Y + EK + + + R DQ
Sbjct 153 VDKEGRPIYIERLGQVDATKLMKVTTIDRYVKYHVKEFEKTFNVKFPACSIAAKRHIDQS 212
Query 208 VFLVDFAGFRVSQV--PSMDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLML 265
++D G +S + D+ + + +I N++Y + L + F+++A +W V+ L
Sbjct 213 TTILDVQGVGLSNFNKAAKDLLQSIQKIDNDNYPETLNRMFIINAGCGFRLLWNTVKSFL 272
Query 266 HPLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGTCEAA 307
P T AK+ + K ++KL + I + L + LGG C A
Sbjct 273 DPKTTAKIHVLGN---KYQTKLLEIIDANELPEFLGGKCTCA 311
> At4g34580
Length=560
Score = 57.8 bits (138), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 59/225 (26%), Positives = 97/225 (43%), Gaps = 32/225 (14%)
Query 104 NDL-ILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHI-------QPEEVMGTAARGSV 155
+DL ++ R+LR+ ++ A+Q +QWR++ I + +EVM +G
Sbjct 84 DDLHMMLRFLRARKFDIEKAKQMWSDMIQWRKDFGADTIIEDFDFEEIDEVMKHYPQGY- 142
Query 156 YRRGFDVHGRPIVYFKPGREPA----QSTRAAQ--DYTLYTLEKALQ------SVPVGSG 203
G D GRP+ + G+ A Q T + Y + EK + SV
Sbjct 143 --HGVDKEGRPVYIERLGQIDANKLLQVTTMDRYVKYHVKEFEKTFKVKFPSCSVAANKH 200
Query 204 RDQVVFLVDFAGFRVSQVPSMDVSREVVQ----ILNEHYTDILAKAFLLDAPGYLDGIWR 259
DQ ++D G + +RE++Q I NE+Y + L + F+++A +W
Sbjct 201 IDQSTTILDVQGVGLKNFSK--SARELLQRLCKIDNENYPETLNRMFIINAGSGFRLLWS 258
Query 260 LVRLMLHPLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGTC 304
V+ L P T AK+ + K SKL + I L + GG C
Sbjct 259 TVKSFLDPKTTAKIHVLGN---KYHSKLLEVIDASELPEFFGGAC 300
> At1g01630
Length=255
Score = 57.0 bits (136), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 49/204 (24%), Positives = 89/204 (43%), Gaps = 12/204 (5%)
Query 104 NDLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPH-HIQPEEVMGTAARGSVYRRGFDV 162
+DL++ R+LR+ + + A + + W+R P HI E+ + + +G D
Sbjct 49 DDLMIRRFLRARDLDIEKASTMFLNYLTWKRSMLPKGHIPEAEIANDLSHNKMCMQGHDK 108
Query 163 HGRPIVYFKPGR-EPAQST-RAAQDYTLYTLEKALQSVPVGSGRDQVVFLVDFAGFRVSQ 220
GRPI R P++ + + +YTLEK +P G+++ V + D G+ S
Sbjct 109 MGRPIAVAIGNRHNPSKGNPDEFKRFVVYTLEKICARMP--RGQEKFVAIGDLQGWGYSN 166
Query 221 VPSMDVSREVVQI--LNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLHPLTAAKVEFINT 278
D+ + + L + Y + L K +++ AP W+++ + T K+ F+
Sbjct 167 C---DIRGYLAALSTLQDCYPERLGKLYIVHAPYIFMTAWKVIYPFIDANTKKKIVFV-- 221
Query 279 SNPKQRSKLTQQIPLHFLEQTLGG 302
N K L + I L GG
Sbjct 222 ENKKLTPTLLEDIDESQLPDIYGG 245
> At3g22410
Length=400
Score = 55.8 bits (133), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 43/183 (23%), Positives = 84/183 (45%), Gaps = 7/183 (3%)
Query 94 LTWEEVLWLSNDLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTAARG 153
LT+++ + + + + R+L+ A +QL + WR+ + EE + G
Sbjct 21 LTFKQEKFCNRECV-ERFLKVKGDNVKKAAKQLSSCLSWRQNFDIERLGAEEFSTELSDG 79
Query 154 SVYRRGFDVHGRPIVYFKPGREPAQSTRAAQDYT---LYTLEKALQSVPVGSGRDQVVFL 210
Y G D RP++ F+ + Q + +T +T+E A+ S+ + V L
Sbjct 80 VAYISGHDRESRPVIIFR-FKHDYQKLHTQKQFTRLVAFTIETAISSMSRNT-EQSFVLL 137
Query 211 VDFAGFRVSQVPSMDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLHPLTA 270
D + FR S + ++ ++I+ ++Y L KAF++D P + +W+ VR + TA
Sbjct 138 FDASFFRSSSAFA-NLLLATLKIIADNYPCRLYKAFIIDPPSFFSYLWKGVRPFVELSTA 196
Query 271 AKV 273
+
Sbjct 197 TMI 199
> At2g21540
Length=371
Score = 55.8 bits (133), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 53/224 (23%), Positives = 96/224 (42%), Gaps = 31/224 (13%)
Query 107 ILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHI-------QPEEVMGTAARGSVYRRG 159
++ R+LR+ ++ A+Q + WR+E I + +EV+ +G G
Sbjct 95 MMLRFLRARKFDLEKAKQMWTDMIHWRKEFGVDTIMEDFDFKEIDEVLKYYPQGY---HG 151
Query 160 FDVHGRPIVYFKPGREPAQSTRAAQ--------DYTLYTLEKALQ------SVPVGSGRD 205
D GRP+ + G+ A T+ Q Y + EK S+ D
Sbjct 152 VDKDGRPVYIERLGQVDA--TKLMQVTTIDRYVKYHVREFEKTFNIKLPACSIAAKKHID 209
Query 206 QVVFLVDF--AGFRVSQVPSMDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRL 263
Q ++D G + + D+ + + +I +++Y + L + F+++A +W V+
Sbjct 210 QSTTILDVQGVGLKSFSKAARDLLQRIQKIDSDNYPETLNRMFIINAGSGFRLLWSTVKS 269
Query 264 MLHPLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGTCEAA 307
L P T AK+ + K +SKL + I + L + LGG C A
Sbjct 270 FLDPKTTAKIHVLGN---KYQSKLLEIIDSNELPEFLGGNCTCA 310
> SPAC589.09
Length=388
Score = 54.7 bits (130), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 55/224 (24%), Positives = 91/224 (40%), Gaps = 34/224 (15%)
Query 103 SNDLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHI-----------QPEEVMGTAA 151
S D IL R+L S N + A Q+L+ T+QWR + I ++ +
Sbjct 78 SPDAILVRFLASCNNDSHEASQKLINTLQWRVDTGVERIVERGELYAKETNDDQFLEQLR 137
Query 152 RGSVYRRGFDVHGRPIVYF-----KPGREPAQSTRAAQDYTLYTLEKALQSVPVGSGRD- 205
G V G D+ RPI Y +P + S R + + T+ L+ P + +D
Sbjct 138 TGKVTMLGRDLSDRPICYIQVHLHQPSKLTQNSLREMTVWVMETMRLFLR--PQKTLKDS 195
Query 206 -----QVVFLVDFAGFRVSQVPSMDVS--REVVQILNEHYTDILAKAFLLDAPGYLDGIW 258
V L D + F + +MD S + + L +Y L L +P +W
Sbjct 196 MDSPQNVNVLFDLSNF---SLHNMDYSFVKYLASCLEYYYPQSLGVCILHKSPWIFRSVW 252
Query 259 RLVRLMLHPLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGG 302
+++ + P AAK+ F ++N L + I + +LGG
Sbjct 253 NIIKGWIKPEIAAKIVFTQSAN-----DLEKYIDYSVIPTSLGG 291
> SPAC3H8.10
Length=286
Score = 54.3 bits (129), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 52/250 (20%), Positives = 108/250 (43%), Gaps = 29/250 (11%)
Query 77 NSSSSSSGSGSKKEVRELTWEEVLWLSNDLILWRYLRSYNWEEAAAQQQLMQTVQWRRE- 135
NS+ ++ + E+++L + E L +D L R+LR+ + + + ++ +WR+E
Sbjct 25 NSTQQATLDSMRLELQKLGYTERL---DDATLLRFLRARKFNLQQSLEMFIKCEKWRKEF 81
Query 136 ------RKPHHIQPEEVMGTAARGSVYRRGFDVHGRPI-------VYFKPGREPAQSTRA 182
+ H+ + E V + Y + D+ GRP+ + K + R
Sbjct 82 GVDDLIKNFHYDEKEAV--SKYYPQFYHKT-DIDGRPVYVEQLGNIDLKKLYQITTPERM 138
Query 183 AQDYTLYTLEKALQSVPVGSGR-----DQVVFLVDFAGFRVSQVPSM-DVSREVVQILNE 236
Q+ AL+ P S + + ++D G ++ + S+ R+ I +
Sbjct 139 MQNLVYEYEMLALKRFPACSRKAGGLIETSCTIMDLKGVGITSIHSVYSYIRQASSISQD 198
Query 237 HYTDILAKAFLLDAPGYLDGIWRLVRLMLHPLTAAKVEFINTSNPKQRSKLTQQIPLHFL 296
+Y + + K ++++AP + L++ L T K+ + ++ +S L +QIP L
Sbjct 199 YYPERMGKFYVINAPWGFSSAFNLIKGFLDEATVKKIHILGSN---YKSALLEQIPADNL 255
Query 297 EQTLGGTCEA 306
LGG C+
Sbjct 256 PAKLGGNCQC 265
> YKL091c
Length=310
Score = 54.3 bits (129), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 50/227 (22%), Positives = 97/227 (42%), Gaps = 34/227 (14%)
Query 104 NDLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHI----------QPEEVMGTAARG 153
+D L R+LR+ ++ A+ + ++T +WR E + I + +E + A
Sbjct 50 DDSTLLRFLRARKFDINASVEMFVETERWREEYGANTIIEDYENNKEAEDKERIKLAKMY 109
Query 154 SVYRRGFDVHGRPIVY-----------FKPGREPAQSTRAAQDYTLYTLEKALQSVPVGS 202
Y D GRP+ + +K E ++Y L+ A VP S
Sbjct 110 PQYYHHVDKDGRPLYFEELGGINLKKMYKITTEKQMLRNLVKEYELF----ATYRVPACS 165
Query 203 GRDQVVF-----LVDFAGFRVSQVPS-MDVSREVVQILNEHYTDILAKAFLLDAPGYLDG 256
R + ++D G +S + ++V I +Y + + K +++ +P
Sbjct 166 RRAGYLIETSCTVLDLKGISLSNAYHVLSYIKDVADISQNYYPERMGKFYIIHSPFGFST 225
Query 257 IWRLVRLMLHPLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGT 303
++++V+ L P+T +K+ + +S K+ L +QIP+ L GGT
Sbjct 226 MFKMVKPFLDPVTVSKIFILGSSYKKE---LLKQIPIENLPVKYGGT 269
> At2g18180
Length=558
Score = 54.3 bits (129), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 49/226 (21%), Positives = 95/226 (42%), Gaps = 31/226 (13%)
Query 107 ILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHI-------QPEEVMGTAARGSVYRRG 159
++ R+L++ ++ Q ++WR+E + + +EV+ +G G
Sbjct 81 MMLRFLKARKFDLEKTNQMWSDMLRWRKEFGADTVMEDFEFKEIDEVLKYYPQG---HHG 137
Query 160 FDVHGRPIVYFKPGREPAQSTRAAQ--------DYTLYTLEKALQ------SVPVGSGRD 205
D GRP+ + G+ ST+ Q +Y + E+ S+ D
Sbjct 138 VDKEGRPVYIERLGQ--VDSTKLMQVTTMDRYVNYHVMEFERTFNVKFPACSIAAKKHID 195
Query 206 QVVFLVDFAGFRVSQV--PSMDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRL 263
Q ++D G + + D+ + ++ ++Y + L + F+++A +W V+
Sbjct 196 QSTTILDVQGVGLKNFNKAARDLITRLQKVDGDNYPETLNRMFIINAGSGFRMLWNTVKS 255
Query 264 MLHPLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGTCEAAYN 309
L P T AK+ + K +SKL + I L + LGG+C A N
Sbjct 256 FLDPKTTAKIHVLGN---KYQSKLLEIIDASELPEFLGGSCTCADN 298
> At4g36490
Length=577
Score = 53.9 bits (128), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 49/225 (21%), Positives = 96/225 (42%), Gaps = 30/225 (13%)
Query 107 ILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHI------QPEEVMGTAARGSVYRRGF 160
++ R+L++ ++ +Q + ++WR+E + + +EV+ +G G
Sbjct 94 MMLRFLKARKFDLEKTKQMWTEMLRWRKEFGADTVMEFDFKEIDEVLKYYPQG---HHGV 150
Query 161 DVHGRPIVYFKPGREPAQSTRAAQ--------DYTLYTLEKALQ------SVPVGSGRDQ 206
D GRP+ + G ST+ Q +Y + E+ S+ DQ
Sbjct 151 DKEGRPVYIERLGL--VDSTKLMQVTTMDRYVNYHVMEFERTFNVKFPACSIAAKKHIDQ 208
Query 207 VVFLVDFAGFRVSQV--PSMDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLM 264
++D G + + D+ + ++ ++Y + L + F+++A +W V+
Sbjct 209 STTILDVQGVGLKNFNKAARDLITRLQKVDGDNYPETLNRMFIINAGSGFRMLWNTVKSF 268
Query 265 LHPLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGTCEAAYN 309
L P T AK+ + K +SKL + I L + LGG+C A N
Sbjct 269 LDPKTTAKIHVLGN---KYQSKLLEIIDESELPEFLGGSCTCADN 310
> CE01099
Length=743
Score = 53.9 bits (128), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 46/223 (20%), Positives = 96/223 (43%), Gaps = 25/223 (11%)
Query 102 LSNDLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTAARGSVYRRGF- 160
L ND L R+LR+ +++ A A+ + ++ WR++ H++ ++++ R +V ++ F
Sbjct 298 LPNDAHLLRFLRARDFDVAKAKDMVHASIIWRKQ---HNV--DKILEEWTRPTVIKQYFP 352
Query 161 ------DVHGRPIVYFKPGREPAQSTRAAQ------DYTLYTLEKALQSVP-----VGSG 203
D GRP+ + G+ + + TL E LQ +G+
Sbjct 353 GCWHNSDKAGRPMYILRFGQLDTKGMLRSCGVENLVKLTLSICEDGLQRAAEATRKLGTP 412
Query 204 RDQVVFLVDFAGFRVSQV--PSMDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLV 261
+VD G + + P + ++++I+ +Y + + + ++ AP +W L+
Sbjct 413 ISSWSLVVDLDGLSMRHLWRPGVQCLLKIIEIVEANYPETMGQVLVVRAPRVFPVLWTLI 472
Query 262 RLMLHPLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGTC 304
+ T K S + +L + I F+ LGG+C
Sbjct 473 SPFIDEKTRKKFMVSGGSGGDLKEELRKHIEEKFIPDFLGGSC 515
> At3g51670
Length=409
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 39/184 (21%), Positives = 85/184 (46%), Gaps = 21/184 (11%)
Query 105 DLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTAARGSV-YRRGFDVH 163
D+IL ++LR+ +++ A + + L + ++WR E K + E++ G V Y RG+D
Sbjct 82 DVILLKFLRARDFKVADSLRMLEKCLEWREEFKAEKLTEEDLGFKDLEGKVAYMRGYDKE 141
Query 164 GRPIVYFKPG--REPAQSTRAAQD---------YTLYTLEKALQSVPVG-SGRDQVVFLV 211
G P+ Y G +E R D + + LE+ ++ + G + ++ +
Sbjct 142 GHPVCYNAYGVFKEKEMYERVFGDEEKLNKFLRWRVQVLERGVKMLHFKPGGVNSIIQVT 201
Query 212 DFAGFRVSQVPSMDV---SREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLHPL 268
D + +P ++ S +++ + ++Y +++A ++ P Y I+ + L
Sbjct 202 D-----LKDMPKRELRVASNQILSLFQDNYPELVATKIFINVPWYFSVIYSMFSPFLTQR 256
Query 269 TAAK 272
T +K
Sbjct 257 TKSK 260
> At2g21520
Length=531
Score = 51.6 bits (122), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 52/220 (23%), Positives = 97/220 (44%), Gaps = 23/220 (10%)
Query 107 ILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHI----QPEEVMGTAARGSVYRRGFDV 162
++ R+L++ ++ A+Q +QWR+E I EE+ G D
Sbjct 14 MMLRFLKARKFDVEKAKQMWADMIQWRKEFGTDTIIQDFDFEEINEVLKHYPQCYHGVDK 73
Query 163 HGRPIVYFKPGR-EPAQSTRAAQ-----DYTLYTLEKALQ----SVPVGSGR--DQVVFL 210
GRPI + G+ +P + + Y + E++ S + + R D +
Sbjct 74 EGRPIYIERLGKVDPNRLMQVTSMDRYVRYHVKEFERSFMIKFPSCTISAKRHIDSSTTI 133
Query 211 VDFAGFRVSQV--PSMDVSREVVQILNEHYTDILAKAFLLDA-PGYLDGIWRLVRLMLHP 267
+D G + + D+ + +I ++Y + L + F+++A PG+ +W V+ L P
Sbjct 134 LDVQGVGLKNFNKSARDLITRLQKIDGDNYPETLHQMFIINAGPGF-RLLWNTVKSFLDP 192
Query 268 LTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGTCEAA 307
T+AK+ + K SKL + I ++ L + LGG C A
Sbjct 193 KTSAKIHVLGY---KYLSKLLEVIDVNELPEFLGGACTCA 229
> At1g30690
Length=540
Score = 51.6 bits (122), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 48/212 (22%), Positives = 93/212 (43%), Gaps = 19/212 (8%)
Query 103 SNDLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTAARGSVYRRGFDV 162
S D+IL ++LR+ +++ A + L +T++WR++ K I EE G + Y G D
Sbjct 218 STDVILLKFLRARDFKVNEAFEMLKKTLKWRKQNKIDSILGEE-FGEDLATAAYMNGVDR 276
Query 163 HGRPIVYFKPGREPAQSTRAAQD------YTLYTLEKALQSVPVGSGRDQVVFLVDFAGF 216
P+ Y E Q+ + ++ + +EK +Q + + G V L+
Sbjct 277 ESHPVCYNVHSEELYQTIGSEKNREKFLRWRFQLMEKGIQKLNLKPG--GVTSLLQIHDL 334
Query 217 R----VSQVPSMDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLHPLTA-- 270
+ VS+ ++V++ L ++Y + +++ ++ P + + R +L P
Sbjct 335 KNAPGVSRTEIWVGIKKVIETLQDNYPEFVSRNIFINVPFWFYAM----RAVLSPFLTQR 390
Query 271 AKVEFINTSNPKQRSKLTQQIPLHFLEQTLGG 302
K +F+ K R L + IP L GG
Sbjct 391 TKSKFVVARPAKVRETLLKYIPADELPVQYGG 422
> YLR380w
Length=408
Score = 51.2 bits (121), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 45/231 (19%), Positives = 93/231 (40%), Gaps = 27/231 (11%)
Query 96 WEEVLWLSNDLILWRYLRSYNWEEAAAQQQLMQTVQWRR----------ERKPHHIQPEE 145
W ++ + D + +++R+ W L + WR+ ER +
Sbjct 100 WHDIKNETPDATILKFIRARKWNADKTIAMLGHDLYWRKDTINKIINGGERAVYENNETG 159
Query 146 VMGTAARGSVYRRGFDVHGRPIVYFKP--GREPAQSTRAAQDYTLYTLEKAL----QSVP 199
V+ +G+D RP++ +P Q+ + + ++L +E++ ++ P
Sbjct 160 VIKNLELQKATIQGYDNDMRPVILVRPRLHHSSDQTEQELEKFSLLVIEQSKLFFKENYP 219
Query 200 VGSGRDQVVFLVDFAGFRVSQVPSMDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWR 259
+ L D GF +S + V + ++ HY + L + AP + IW
Sbjct 220 AST-----TILFDLNGFSMSNMDYAPV-KFLITCFEAHYPESLGHLLIHKAPWIFNPIWN 273
Query 260 LVRLMLHPLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGTCEAAYNH 310
+++ L P+ A+K+ F T N + K Q ++ + LGG + +H
Sbjct 274 IIKNWLDPVVASKIVF--TKNIDELHKFIQP---QYIPRYLGGENDNDLDH 319
> At1g75370
Length=530
Score = 49.7 bits (117), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 48/218 (22%), Positives = 90/218 (41%), Gaps = 21/218 (9%)
Query 108 LWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTAARGSVYR----RGFDVH 163
+ R+L++ ++ + ++WR++ I + Y G D
Sbjct 1 MLRFLKARKFDIGKTKLMWSNMIKWRKDFGTDTIFEDFEFEEFDEVLKYYPHGYHGVDKE 60
Query 164 GRPIVYFKPGR-EPAQSTRAAQ-----DYTLYTLEKA----LQSVPVGSGR--DQVVFLV 211
GRP+ + G +PA+ + Y + EK L + + + R D ++
Sbjct 61 GRPVYIERLGLVDPAKLMQVTTVERFIRYHVREFEKTVNIKLPACCIAAKRHIDSSTTIL 120
Query 212 DF--AGFRVSQVPSMDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLHPLT 269
D GF+ P+ D+ ++ +I N++Y + L + F+++ +W V+ L P T
Sbjct 121 DVQGVGFKNFSKPARDLIIQLQKIDNDNYPETLHRMFIINGGSGFKLVWATVKQFLDPKT 180
Query 270 AAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGTCEAA 307
K+ I K ++KL + I L LGGTC A
Sbjct 181 VTKIHVIGN---KYQNKLLEIIDASQLPDFLGGTCTCA 215
> At1g14820
Length=252
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 49/200 (24%), Positives = 84/200 (42%), Gaps = 8/200 (4%)
Query 108 LWRYLRSYNWEEAAAQQQLMQTVQWRRERKP--HHIQPEEVMGTAARGSVYRRGFDVHGR 165
L R+L + + + A + + +WR P I EV V +G G
Sbjct 31 LMRFLVARSMDPVKAAKMFVDWQKWRASMVPPTGFIPESEVQDELEFRKVCLQGPTKSGH 90
Query 166 PIVYFKPGREPAQSTRAA-QDYTLYTLEKALQSVPVGS--GRDQVVFLVDFAGFRVSQVP 222
P+V + A A + + +Y L+K + S G G +++V ++D A +
Sbjct 91 PLVLVITSKHFASKDPANFKKFVVYALDKTIASGNNGKEVGGEKLVAVIDLANITYKNLD 150
Query 223 SMDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLHPLTAAKVEFINTSNPK 282
+ + Q L +Y + LAK ++L PG+ +W+ V L T K+ I T +
Sbjct 151 ARGLITGF-QFLQSYYPERLAKCYILHMPGFFVTVWKFVCRFLEKATQEKI-VIVTDGEE 208
Query 283 QRSKLTQQIPLHFLEQTLGG 302
QR K ++I L + GG
Sbjct 209 QR-KFEEEIGADALPEEYGG 227
> At1g19650
Length=616
Score = 48.9 bits (115), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 48/222 (21%), Positives = 95/222 (42%), Gaps = 27/222 (12%)
Query 107 ILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHI-------QPEEVMGTAARGSVYRRG 159
I+ R+L + ++ A+ +QWRR+ I + +EV+ +G G
Sbjct 104 IMLRFLFARKFDLGKAKLMWTNMIQWRRDFGTDTILEDFEFPELDEVLRYYPQGY---HG 160
Query 160 FDVHGRPIVYFKPGREPAQS---TRAAQDYTLYTLEKALQSVPV-------GSGR--DQV 207
D GRP+ + G+ A + Y Y +++ +++ V + R D
Sbjct 161 VDKEGRPVYIERLGKVDASKLMQVTTLERYLRYHVKEFEKTITVKFPACCIAAKRHIDSS 220
Query 208 VFLVDFAGFRVSQVP--SMDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLML 265
++D G + + D+ ++ +I +++Y + L + F+++A +W V+ L
Sbjct 221 TTILDVQGLGLKNFTKTARDLIIQLQKIDSDNYPETLHRMFIINAGSGFKLLWGTVKSFL 280
Query 266 HPLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGTCEAA 307
P T +K+ + K ++KL + I L GGTC A
Sbjct 281 DPKTVSKIHVLGN---KYQNKLLEMIDASQLPDFFGGTCTCA 319
> At2g16380
Length=582
Score = 48.1 bits (113), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 52/224 (23%), Positives = 96/224 (42%), Gaps = 24/224 (10%)
Query 104 NDL-ILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHI----QPEEVMGTAARGSVYRR 158
+DL ++ R+LR+ +++ A+Q +QWR + I + EE+
Sbjct 82 DDLHMMLRFLRARKFDKEKAKQMWSDMLQWRMDFGVDTIIEDFEFEEIDQVLKHYPQGYH 141
Query 159 GFDVHGRPIVYFKPGREPAQSTRAA------QDYTLYTLEKALQ------SVPVGSGRDQ 206
G D GRP+ + G+ A A + Y + EK + S DQ
Sbjct 142 GVDKEGRPVYIERLGQIDANKLLQATTMDRYEKYHVKEFEKMFKIKFPSCSAAAKKHIDQ 201
Query 207 --VVFLVDFAGFRVSQVPSMDVSREVVQILNEHYTDILAKAFLLDA-PGYLDGIWRLVRL 263
+F V G + + ++ + +++I N++Y + L + F+++A PG+ +W ++
Sbjct 202 STTIFDVQGVGLKNFNKSARELLQRLLKIDNDNYPETLNRMFIINAGPGF-RLLWAPIKK 260
Query 264 MLHPLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGTCEAA 307
L P T +K+ + K + KL + I L GG C A
Sbjct 261 FLDPKTTSKIHVLGN---KYQPKLLEAIDASELPYFFGGLCTCA 301
> YMR079w
Length=304
Score = 47.8 bits (112), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 50/220 (22%), Positives = 87/220 (39%), Gaps = 20/220 (9%)
Query 104 NDLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHI----QPEEVMGTAARGSVYRRG 159
+D L R+LR+ ++ A++ +WR++ I +E A Y
Sbjct 54 DDSTLLRFLRARKFDVQLAKEMFENCEKWRKDYGTDTILQDFHYDEKPLIAKFYPQYYHK 113
Query 160 FDVHGRPIVYFKPG------REPAQSTRAAQDYTLYTLEKALQ------SVPVGSGRDQV 207
D GRP+ + + G S ++ E +Q S G +
Sbjct 114 TDKDGRPVYFEELGAVNLHEMNKVTSEERMLKNLVWEYESVVQYRLPACSRAAGHLVETS 173
Query 208 VFLVDFAGFRVSQVPS-MDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLH 266
++D G +S S M RE I +Y + + K ++++AP +RL + L
Sbjct 174 CTIMDLKGISISSAYSVMSYVREASYISQNYYPERMGKFYIINAPFGFSTAFRLFKPFLD 233
Query 267 PLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGTCEA 306
P+T +K+ + +S K+ L +QIP L GG E
Sbjct 234 PVTVSKIFILGSSYQKE---LLKQIPAENLPVKFGGKSEV 270
> At1g22530
Length=683
Score = 47.8 bits (112), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 37/66 (56%), Gaps = 0/66 (0%)
Query 104 NDLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTAARGSVYRRGFDVH 163
+D+IL ++LR+ +++ A L TVQWR+E K + E++ G+ V+ G D
Sbjct 364 SDVILLKFLRARDFKVKEAFTMLKNTVQWRKENKIDDLVSEDLEGSEFEKLVFTHGVDKQ 423
Query 164 GRPIVY 169
G ++Y
Sbjct 424 GHVVIY 429
> At1g72160
Length=490
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 37/187 (19%), Positives = 86/187 (45%), Gaps = 18/187 (9%)
Query 104 NDLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTAARGSVYRRGFDVH 163
+D++L ++LR+ ++ + L T++WR+E K + E+++ + V+ G D
Sbjct 163 SDVVLLKFLRAREFKVKDSFAMLKNTIKWRKEFKIDELVEEDLVDDLDK-VVFMHGHDRE 221
Query 164 GRPIVYFKPGR-------EPAQSTRAAQDYTLYT----LEKALQSVPVGSGRDQVVFLVD 212
G P+ Y G S + + L T LE++++ + SG +F V+
Sbjct 222 GHPVCYNVYGEFQNKELYNKTFSDEEKRKHFLRTRIQFLERSIRKLDFSSGGVSTIFQVN 281
Query 213 FA----GFRVSQVPSMDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLHPL 268
G ++ S +++ V++L ++Y + + K ++ P + + ++ + P
Sbjct 282 DMKNSPGLGKKELRS--ATKQAVELLQDNYPEFVFKQAFINVPWWYLVFYTVIGPFMTPR 339
Query 269 TAAKVEF 275
+ +K+ F
Sbjct 340 SKSKLVF 346
> YJL145w
Length=294
Score = 44.7 bits (104), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 37/183 (20%), Positives = 80/183 (43%), Gaps = 19/183 (10%)
Query 105 DLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTA----------ARGS 154
D + ++ ++Y +E + Q L+ + WRRE P +EV T A G
Sbjct 58 DRLTYKLCKAYQFEYSTIVQNLIDILNWRREFNPLSCAYKEVHNTELQNVGILTFDANGD 117
Query 155 VYRRG--FDVHGRPIVYFKPGREPAQSTRAAQDYTLYTLEKALQSVPVGSGRDQVVFLV- 211
++ ++++G+ + +E Q+ Y + +EK L + S + + V
Sbjct 118 ANKKAVTWNLYGQLV----KKKELFQNVDKFVRYRIGLMEKGLSLLDFTSSDNNYMTQVH 173
Query 212 DFAGFRVSQVPS--MDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLHPLT 269
D+ G V ++ S + S+ V+ I ++Y ++L + ++ P ++ L++ + T
Sbjct 174 DYKGVSVWRMDSDIKNCSKTVIGIFQKYYPELLYAKYFVNVPTVFGWVYDLIKKFVDETT 233
Query 270 AAK 272
K
Sbjct 234 RKK 236
> At5g56160
Length=592
Score = 44.3 bits (103), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 48/222 (21%), Positives = 96/222 (43%), Gaps = 28/222 (12%)
Query 107 ILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHI-------QPEEVMGTAARGSVYRRG 159
+L R+L++ ++ + ++WR+E I + +EV +G G
Sbjct 109 MLLRFLKTMEFKIEKTVTAWEEMLKWRKEFGTDRIIQDFNFKELDEVTRHYPQGY---HG 165
Query 160 FDVHGRPIVYFKPGR-EPAQ--STRAAQDYTLY-------TLEKALQSVPVGSGR----D 205
D GRPI + G+ P + + Y Y TL++ L + V + R
Sbjct 166 VDKDGRPIYIERLGKAHPGKLMEVTTIERYLKYHVQEFERTLQEKLPACSVAAKRRVTTT 225
Query 206 QVVFLVDFAGFRVSQVPSMDVSREVVQILNEHYTDILAKAFLLDAP-GYLDGIWRLVRLM 264
+ V+ G + + ++ + ++ +Y + L + F+++A G+ +W + +
Sbjct 226 TTILDVEGLGMKNFTPTAANLLATIAKVDCNYYPETLHRMFIVNAGIGFRSFLWPAAQKL 285
Query 265 LHPLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGTCEA 306
L P+T AK++ + P+ SKL + I L + LGG C+
Sbjct 286 LDPMTIAKIQVLE---PRSLSKLLEAIDSSQLPEFLGGLCKC 324
> Hs4506301
Length=593
Score = 43.1 bits (100), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/115 (23%), Positives = 57/115 (49%), Gaps = 9/115 (7%)
Query 188 LYTLEKALQSVPVGSGRDQVVFLVDFAGFRVSQVPSMDVSREVVQILNEHYTDILAKAFL 247
Y L++A+ S + R+ +VF+ D G + +D+ ++V+ +L + L K +
Sbjct 132 FYLLDRAVDSFE--TQRNGLVFIYDMCGSNYANF-ELDLGKKVLNLLKGAFPARLKKVLI 188
Query 248 LDAPGYLDGIWRLVRLMLHPLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGG 302
+ AP + + ++ L+L +++ + TS ++TQ +P L + LGG
Sbjct 189 VGAPIWFRVPYSIISLLLKDKVRERIQILKTS------EVTQHLPRECLPENLGG 237
> CE20032
Length=368
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 40/218 (18%), Positives = 92/218 (42%), Gaps = 29/218 (13%)
Query 110 RYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTAARGS-------VYRRGFDV 162
R+L + N + +Q+++Q + R+ I ++ T G + + ++V
Sbjct 41 RFLNANNGDVEKTRQKIIQLFEHRKLMGYEKINDLDIFTTVNIGKDCFERFHISQLNYEV 100
Query 163 HGRPIVYFKPGREPAQSTRAAQ----DYTLYT-------LEKALQSVPVGSGR-DQVVFL 210
+ + F E + Y L++ +A+ +G+ VV +
Sbjct 101 TSKNLHVFVQKMEGTDIKEILKVMPLSYVLHSYFMLQENFSRAMAHTERKTGKTSSVVCI 160
Query 211 VDFAGFRVSQVPS-----MDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLML 265
+D G + + ++R VVQ+ E++++ L K L++ PG + +W++ + ++
Sbjct 161 LDLKGLNLMDFMNPLSGPAQLARLVVQVWAEYFSEHLCKLLLINPPGIISVMWQVTKRLV 220
Query 266 HPLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGT 303
P T K+ F++ ++ + IP+ + GGT
Sbjct 221 DPNTVEKLAFLSNVEDLKKYLEPEAIPVEY-----GGT 253
> SPBC365.01
Length=355
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 50/212 (23%), Positives = 89/212 (41%), Gaps = 18/212 (8%)
Query 96 WEEVLWLSNDLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMG---TAAR 152
+E+ + DL L R+L++ + + L + WR++ I G +
Sbjct 43 FEQCAFDDFDLTLLRFLKARKFVVTDSSDMLANAIVWRQQANLRSIMVRGENGLNQNFVK 102
Query 153 GSVYRR-GFDVHGRPIVYFK-PGREPAQSTRAAQD---YTLYTLEKA-LQSVPVGSGRDQ 206
S+Y G D GR IV+ P ++T+ ++ LY +E A L +
Sbjct 103 ASMYFIWGQDKKGRAIVFLNLHNFIPPKNTKDMEELKALILYAMENARLFLDSEQNAAKG 162
Query 207 VVFLVDFAGFRVSQVPSMDVSREVVQILNEHYTDILAKAFLLDAP---GYLDGIWRLVRL 263
V+ LVD F + +D +R + +Y +IL +A ++ + +G+W + +
Sbjct 163 VLGLVDLTYFSRKNI-DLDFARVFAETFQNYYPEILGQALIVGSGFRMALFEGVWSIGKY 221
Query 264 MLHPLTAAKVEFINTSNPKQRSKL--TQQIPL 293
L P +KV F P Q S ++ IPL
Sbjct 222 FLDPEVRSKVTF---CKPAQVSGYVDSKYIPL 250
> Hs4506867
Length=715
Score = 41.6 bits (96), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 43/214 (20%), Positives = 83/214 (38%), Gaps = 19/214 (8%)
Query 108 LWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHI----QPEEVMGTAARGSVYRRGFDVH 163
+ R+LR+ ++ A++ + Q++ WR++ + +I P +V+ G + D
Sbjct 280 ILRFLRARDFNIDKAREIMCQSLTWRKQHQVDYILETWTPPQVLQDYYAGGWHHH--DKD 337
Query 164 GRPIVYFKPGREP------AQSTRAAQDYTLYTLEKALQSVP-----VGSGRDQVVFLVD 212
GRP+ + G+ A A Y L E+ L+ G LVD
Sbjct 338 GRPLYVLRLGQMDTKGLVRALGEEALLRYVLSVNEERLRRCEENTKVFGRPISSWTCLVD 397
Query 213 FAGFRVSQV--PSMDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLHPLTA 270
G + + P + +++++ +Y + L + +L AP +W LV + T
Sbjct 398 LEGLNMRHLWRPGVKALLRIIEVVEANYPETLGRLLILRAPRVFPVLWTLVSPFIDDNTR 457
Query 271 AKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGTC 304
K ++ + L I + L G C
Sbjct 458 RKFLIYAGNDYQGPGGLLDYIDKEIIPDFLSGEC 491
> At5g47730
Length=341
Score = 41.6 bits (96), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 42/205 (20%), Positives = 87/205 (42%), Gaps = 19/205 (9%)
Query 108 LWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTA----ARGS--VYRRGFD 161
L R+L++ +W A L++ ++WR + + I + ++ T R S + G+
Sbjct 39 LGRFLKARDWNVCKAHTMLVECLRWRVDNEIDSILSKPIVPTELYRDVRDSQLIGMSGYT 98
Query 162 VHGRPIVYFKPGREPAQSTRAAQDYTL--------YTLEKALQSVPVGSGR--DQVVFLV 211
G P+ F G + +A+ Y + Y L S+ +GR V ++
Sbjct 99 KEGLPV--FAIGVGLSTFDKASVHYYVQSHIQINEYRDRVLLPSISKKNGRPITTCVKVL 156
Query 212 DFAGFRVSQVPSMDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLHPLTAA 271
D G ++S + + + + I + +Y + ++++AP W++V+ +L T
Sbjct 157 DMTGLKLSALSQIKLVTIISTIDDLNYPEKTNTYYVVNAPYIFSACWKVVKPLLQERTRK 216
Query 272 KVEFINTSNPKQRSKLTQQIPL-HF 295
KV ++ + K+ L HF
Sbjct 217 KVHVLSGCGRDELLKIMDFTSLPHF 241
> Hs14779875
Length=696
Score = 41.2 bits (95), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 41/218 (18%), Positives = 93/218 (42%), Gaps = 19/218 (8%)
Query 102 LSNDLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHI----QPEEVMGTAARGSVYR 157
+ D + R+LR++++ A++ L Q++ WR++ + + QP ++ G +
Sbjct 261 IPKDEHILRFLRAHDFHLDKAREMLRQSLSWRKQHQVDLLLQTWQPPALLEEFYAGGWHY 320
Query 158 RGFDVHGRPIVYFKPGREPAQS-TRAAQDYTL----YTLEKALQSVPVGSGR------DQ 206
+ D+ GRP+ + G+ + +A + L ++ + Q GS R
Sbjct 321 Q--DIDGRPLYILRLGQMDTKGLMKAVGEEALLRHVLSVNEEGQKRCEGSTRQLGRPISS 378
Query 207 VVFLVDFAGFRVSQV--PSMDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLM 264
L+D G + + P + +++++ ++Y + L + ++ AP +W L+
Sbjct 379 WTCLLDLEGLNMRHLWRPGVKALLRMIEVVEDNYPETLGRLLIVRAPRVFPVLWTLISPF 438
Query 265 LHPLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGG 302
++ T K + SN + L + + LGG
Sbjct 439 INENTRRKFLIYSGSNYQGPGGLVDYLDREVIPDFLGG 476
> At4g09160
Length=723
Score = 41.2 bits (95), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 34/186 (18%), Positives = 87/186 (46%), Gaps = 18/186 (9%)
Query 105 DLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTAARGSVYRRGFDVHG 164
D++L ++LR+ +++ A L +T+QWR + + +E +G V+ +G D
Sbjct 339 DVVLLKFLRARDFKPQEAYSMLNKTLQWRIDFNIEELL-DENLGDDLDKVVFMQGQDKEN 397
Query 165 RPIVYFKPG----REPAQSTRAAQD-------YTLYTLEKALQSVPVGSGRDQVVFLVDF 213
P+ Y G ++ Q T + ++ + + LEK+++++ +G + V+
Sbjct 398 HPVCYNVYGEFQNKDLYQKTFSDEEKRERFLRWRIQFLEKSIRNLDFVAGGVSTICQVN- 456
Query 214 AGFRVSQVPSMD----VSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLHPLT 269
+ S P +++ + +L ++Y + ++K ++ P + +R++ + +
Sbjct 457 -DLKNSPGPGKTELRLATKQALHLLQDNYPEFVSKQIFINVPWWYLAFYRIISPFMSQRS 515
Query 270 AAKVEF 275
+K+ F
Sbjct 516 KSKLVF 521
> At5g47510
Length=403
Score = 40.8 bits (94), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 47/220 (21%), Positives = 90/220 (40%), Gaps = 25/220 (11%)
Query 108 LWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPE---EVMGTAARGSVYRRGF---D 161
L R+L+ +++ +++ + ++WR + K I + E G + Y GF D
Sbjct 51 LRRFLKMRDFDLEKSKEAFLNYMKWRVDYKVDLISQKFKFEEYGEVKKH--YPHGFHKVD 108
Query 162 VHGRPIVYFKPGREPAQSTRAAQ------DYTLYTLEKALQ------SVPVGSGRDQVVF 209
GRPI + G + A +Y + EK + S+
Sbjct 109 KTGRPIYIERLGMTDLNAFLKATTIERYVNYHIKEQEKTMSLRYPACSIASDKHVSSTTT 168
Query 210 LVDFAGFRVSQV--PSMDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLHP 267
++D +G +S P+ + E+ +I + +Y + L + F+++A +W ++ L
Sbjct 169 ILDVSGVGMSNFSKPARSLFMEIQKIDSNYYPETLHRLFVVNASSGFRMLWLALKTFLDA 228
Query 268 LTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGTCEAA 307
T AKV+ + P +L + I L LGG C +
Sbjct 229 RTLAKVQVL---GPNYLGELLEAIEPSNLPTFLGGNCTCS 265
> 7304163
Length=482
Score = 40.0 bits (92), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 37/134 (27%), Positives = 58/134 (43%), Gaps = 17/134 (12%)
Query 125 QLMQTVQWRRERKPHHIQPEEVMGTAARGSVYRRGFDVHGRPIVYFKPGREPAQSTRAAQ 184
+LM++ + R ER Q E++ + G +Y+ G D GRP++ F PAQ+
Sbjct 303 ELMESPEERYERLLRRAQVEDLTEVSGIGCLYQSGVDRLGRPVIVFCGKWFPAQNI---- 358
Query 185 DYTLYTLEKALQSV-----PVGSGRDQVVFLVDFAGFRVSQVPSMDVSREVVQILNEHYT 239
LEKAL + P+ G + + + PS+ REV +L Y
Sbjct 359 -----DLEKALLYLIKLLDPIVKGDYVISYFHTLTS--TNNYPSLHWLREVYSVLPYKYK 411
Query 240 DILAKAFLLDAPGY 253
L KAF + P +
Sbjct 412 KNL-KAFYIVHPTF 424
> At3g46450
Length=486
Score = 40.0 bits (92), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 42/203 (20%), Positives = 84/203 (41%), Gaps = 15/203 (7%)
Query 108 LWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTAARGSVYRRGFDVHGRPI 167
L R+ R N + + + +T+ WR + + I EE + T + ++ G+D + RP
Sbjct 207 LHRFYRVSNGDFTSLLSSIKKTIHWR---ETYRILSEEELETWS-SLLFWHGYDKNQRPC 262
Query 168 VYFKPG----REPAQSTRAAQDYTLYTLEKALQSVPVGSGRDQVVFLVDFAGFRVSQVPS 223
+ + G + P+ + +E + + + ++ LVD G ++P
Sbjct 263 LIVRLGLAFLKLPSHERPRFAQAIISQVEHGVLHL-LTPENSELTVLVDCEGLSPLRIP- 320
Query 224 MDVSREVVQILNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLHPLTAAKVEFINTSNPKQ 283
M + R IL +H+ + L F++ P + I + +L P T K+ + +
Sbjct 321 MQMMRSCSSILQDHFPNRLGCLFIIRLPPVVRVISQTFIQILRPTTRKKLRIEGETFHRV 380
Query 284 RSKLTQQIPLHFLEQTLGGTCEA 306
S+ Q +P + LG C
Sbjct 381 LSEYLQTLPSY-----LGSNCNC 398
> At1g72150
Length=573
Score = 38.5 bits (88), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 33/66 (50%), Gaps = 0/66 (0%)
Query 104 NDLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTAARGSVYRRGFDVH 163
+D+IL ++LR+ +++ A L TVQWR+E K + + V+ G D
Sbjct 255 SDVILTKFLRARDFKVKEALTMLKNTVQWRKENKIDELVESGEEVSEFEKMVFAHGVDKE 314
Query 164 GRPIVY 169
G ++Y
Sbjct 315 GHVVIY 320
> At1g55690
Length=648
Score = 37.7 bits (86), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 46/218 (21%), Positives = 88/218 (40%), Gaps = 22/218 (10%)
Query 108 LWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTAARGSVYR----RGFDVH 163
L R+L++ + Q + ++WR+E I + Y G D
Sbjct 129 LLRFLKARDLNIEKTTQLWEEMLRWRKEYGTDTILEDFDFEELEEVLQYYPQGYHGVDKE 188
Query 164 GRPIVYFKPGR-EPAQSTRAAQ-----DYTLYTLEKALQ----SVPVGSGRD--QVVFLV 211
GRP+ + G+ P++ R Y + E+ALQ + + + R ++
Sbjct 189 GRPVYIERLGKAHPSKLMRITTIDRYLKYHVQEFERALQEKFPACSIAAKRRICSTTTIL 248
Query 212 DFAGFRVSQVP--SMDVSREVVQILNEHYTDILAKAFLLDA-PGYLDGIWRLVRLMLHPL 268
D G + + ++ + +I N +Y + L + ++++A G+ +W + L
Sbjct 249 DVQGLGIKNFTPTAANLVAAMSKIDNSYYPETLHRMYIVNAGTGFKKMLWPAAQKFLDAK 308
Query 269 TAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGTCEA 306
T AK+ + PK KL + I L + LGG+C
Sbjct 309 TIAKIHVLE---PKSLFKLHEVIDSSQLPEFLGGSCSC 343
> At1g05370
Length=439
Score = 37.0 bits (84), Expect = 0.082, Method: Compositional matrix adjust.
Identities = 27/113 (23%), Positives = 44/113 (38%), Gaps = 4/113 (3%)
Query 102 LSNDLILWRYLRSYNWEEAAAQQQLMQTVQWRRERKPHHIQPEEVMGTAARGSVYRRGFD 161
N + R+LR A +QL + WR + +E A G Y G D
Sbjct 39 FCNRACVGRFLRIKGDNVKKAAKQLRSCLSWRSSLGIESLIADEFTAELAEGLAYVAGLD 98
Query 162 VHGRPIVYF--KPGREPAQSTRAAQDYTLYTLEKALQSVPVGSGRDQVVFLVD 212
RP++ F K + + + ++TLE A+ + + +Q V L D
Sbjct 99 DECRPVLVFRIKQDYQKLHTQKQLTRLVVFTLEVAIST--MSRNVEQFVILFD 149
> At1g03280
Length=506
Score = 36.2 bits (82), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 32/120 (26%), Positives = 54/120 (45%), Gaps = 22/120 (18%)
Query 9 VYGYGPPPSLGVHPPIETVEGLNLSSDQLQKVRALRAVVDALPIVEELQQEAPVAKEKSW 68
V G SL V PP EG+NL+ +Q E++QEA V
Sbjct 321 VKSKGGDSSLKVLPPWMIKEGMNLTEEQRG----------------EMRQEAKVDGGAGA 364
Query 69 LSGIFSRSNSSSSSSGSGSKKEVRELTWEEVL--WLSNDLILWRYLRSYNWEEAAAQQQL 126
+ + S+ S+ G+G +K+++ W ++L +L + YL++Y + E QQ+L
Sbjct 365 AAKL---SDDKKSAIGNGDEKDLKACFWGQILYIYLDSHSSFDEYLKAY-YAELMKQQEL 420
> CE28959
Length=408
Score = 35.8 bits (81), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 27/113 (23%), Positives = 53/113 (46%), Gaps = 10/113 (8%)
Query 196 QSVPVGSGRDQVVFLVDFAGFRVSQV--PSMDVSREVVQILNEHYTDILAKAFLLDAPGY 253
Q P+G+ V+F D G + Q+ ++ V ++ L E + D++ K F+++ P +
Sbjct 164 QGKPLGTS---VIF--DLDGLSMVQIDLAALKVVTTMLSQLQEMFPDVIRKIFIVNTPTF 218
Query 254 LDGIWRLVRLMLHPLTAAKVEFINTSNPKQRSKLTQQIPLHFLEQTLGGTCEA 306
+ +W ++ L T KV+ + + L + I L + GGT +A
Sbjct 219 IQVLWSMISPCLAKQTQQKVKILGND---WKQHLKENIGEEVLFERWGGTRKA 268
> CE09506
Length=377
Score = 34.7 bits (78), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 42/229 (18%), Positives = 91/229 (39%), Gaps = 40/229 (17%)
Query 79 SSSSSGSGSKKEVRELTWEEVL-WLSNDLILWRYLRSYNWEEAAAQQQLMQTVQWR---- 133
S S S + E+RE E + + D L R+L+ ++++ + +L+ + +R
Sbjct 2 SISESDRVAINELREAVKEHLTPYYDTDFNLLRWLKGHDYKMDVIKPKLINHLLFRKSDW 61
Query 134 ---------RERKPHH-----------IQPEEVMGTAARGSVYRRGFDVHGRPIVYFKPG 173
R+ HH I P ++ GS G +H P
Sbjct 62 DLDSLADKPRDHPVHHHWKTGLTGESGIIPNTIVNIEQTGSNDYWGM-LHSYP------- 113
Query 174 REPAQSTRAAQDYTLYTLEKALQSVPVGSGRD-QVVFLVDFAGFRVSQVPSMDVSREVVQ 232
A+ + L ++ KA+ + + + V++++D G + + ++ +
Sbjct 114 ---TNEILRARVHDLESMLKAVMDLEKKTNQQCSVIYIMDLTGIKFDKRTITLLTGGLSA 170
Query 233 I---LNEHYTDILAKAFLLDAPGYLDGIWRLVRLMLHPLTAAKVEFINT 278
I + EHY +++ L++ P ++ IW + + +L T K +N+
Sbjct 171 ISAFMAEHYVELVHSFVLVNVPAFISAIWTIAKPLLPERTRNKCNILNS 219
Lambda K H
0.315 0.129 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9716962222
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40