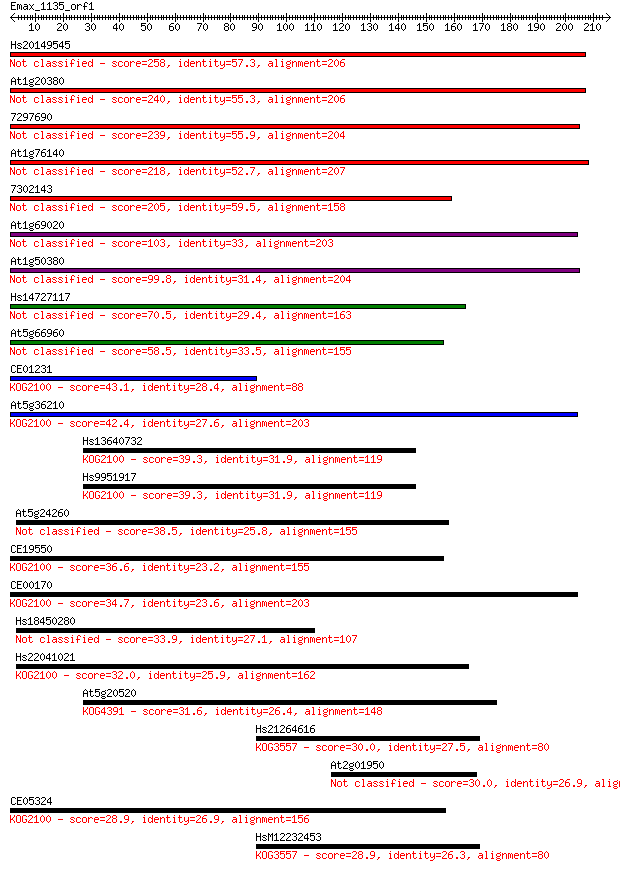
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1135_orf1
Length=215
Score E
Sequences producing significant alignments: (Bits) Value
Hs20149545 258 6e-69
At1g20380 240 2e-63
7297690 239 2e-63
At1g76140 218 5e-57
7302143 205 5e-53
At1g69020 103 2e-22
At1g50380 99.8 3e-21
Hs14727117 70.5 3e-12
At5g66960 58.5 9e-09
CE01231 43.1 4e-04
At5g36210 42.4 7e-04
Hs13640732 39.3 0.006
Hs9951917 39.3 0.006
At5g24260 38.5 0.011
CE19550 36.6 0.042
CE00170 34.7 0.15
Hs18450280 33.9 0.26
Hs22041021 32.0 1.1
At5g20520 31.6 1.2
Hs21264616 30.0 3.4
At2g01950 30.0 3.7
CE05324 28.9 7.4
HsM12232453 28.9 8.6
> Hs20149545
Length=710
Score = 258 bits (659), Expect = 6e-69, Method: Compositional matrix adjust.
Identities = 118/206 (57%), Positives = 152/206 (73%), Gaps = 0/206 (0%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
NIRGGGEYG W+K I NKQ +DDFQ AAEYL +GY++P +L I GGSNGGLLV A
Sbjct 503 NIRGGGEYGETWHKGGILANKQNCFDDFQCAAEYLIKEGYTSPKRLTINGGSNGGLLVAA 562
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDYGNPDKEEDFKYIYKISPIHNINP 120
CANQRPDLF V+A+VGVMD+L+F+K+TIGHAW +DYG D ++ F+++ K SP+HN+
Sbjct 563 CANQRPDLFGCVIAQVGVMDMLKFHKYTIGHAWTTDYGCSDSKQHFEWLVKYSPLHNVKL 622
Query 121 EVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYSSPVSTPLIAHIDTKAGHGG 180
+ P++L++TADHDDRV P HSLK+IA LQ+ VG S S PL+ H+DTKAGHG
Sbjct 623 PEADDIQYPSMLLLTADHDDRVVPLHSLKFIATLQYIVGRSRKQSNPLLIHVDTKAGHGA 682
Query 181 GKPLMKVLDEQALTYGFIASVLGLKW 206
GKP KV++E + + FIA L + W
Sbjct 683 GKPTAKVIEEVSDMFAFIARCLNVDW 708
> At1g20380
Length=731
Score = 240 bits (612), Expect = 2e-63, Method: Compositional matrix adjust.
Identities = 114/213 (53%), Positives = 151/213 (70%), Gaps = 7/213 (3%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
NIRGGGEYG W+K+ NKQ +DDF S AEYL + GY+ P +L I GGSNGG+LVGA
Sbjct 517 NIRGGGEYGEEWHKSGALANKQNCFDDFISGAEYLVSAGYTQPRKLCIEGGSNGGILVGA 576
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDYGNPDKEEDFKYIYKISPIHNI-N 119
C NQRPDLF +A VGVMD+LRF+KFTIGHAW S++G DKEE+F ++ K SP+HN+
Sbjct 577 CINQRPDLFGCALAHVGVMDMLRFHKFTIGHAWTSEFGCSDKEEEFHWLIKYSPLHNVKR 636
Query 120 PEVVRSD---KQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYS---SPVSTPLIAHID 173
P ++D + P+ +++TADHDDRV P HS K +A +Q+ +G S SP + P+IA I+
Sbjct 637 PWEQKTDLFFQYPSTMLLTADHDDRVVPLHSYKLLATMQYELGLSLENSPQTNPIIARIE 696
Query 174 TKAGHGGGKPLMKVLDEQALTYGFIASVLGLKW 206
KAGHG G+P K++DE A Y F+A ++ W
Sbjct 697 VKAGHGAGRPTQKMIDEAADRYSFMAKMVDASW 729
> 7297690
Length=717
Score = 239 bits (611), Expect = 2e-63, Method: Compositional matrix adjust.
Identities = 114/204 (55%), Positives = 143/204 (70%), Gaps = 0/204 (0%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
N+RGGGEYG W+ NKQ +DDFQ+AAEYL Y+T D+LAI GGSNGGLLVG+
Sbjct 509 NLRGGGEYGEKWHNGGRLLNKQNVFDDFQAAAEYLIENKYTTKDRLAIQGGSNGGLLVGS 568
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDYGNPDKEEDFKYIYKISPIHNINP 120
C NQRPDLF A VA+VGVMD+LRF+KFTIGHAW SDYGNP ++E F +YK SP+HN++
Sbjct 569 CINQRPDLFGAAVAQVGVMDMLRFHKFTIGHAWCSDYGNPSEKEHFDNLYKFSPLHNVHT 628
Query 121 EVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYSSPVSTPLIAHIDTKAGHGG 180
+ P+ L++TADHDDRVSP HSLK+IA LQ AV S P++ + KAGHG
Sbjct 629 PKGAETEYPSTLILTADHDDRVSPLHSLKFIAALQEAVRDSEFQKNPVLLRVYQKAGHGA 688
Query 181 GKPLMKVLDEQALTYGFIASVLGL 204
GKP K ++E F++ L +
Sbjct 689 GKPTSKRIEEATDILTFLSKSLNV 712
> At1g76140
Length=724
Score = 218 bits (556), Expect = 5e-57, Method: Compositional matrix adjust.
Identities = 109/214 (50%), Positives = 142/214 (66%), Gaps = 14/214 (6%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
NIRGGGEYG W+KA KQ +DDF S AEYL + GY+ P +L I GGSNGGLL
Sbjct 517 NIRGGGEYGEEWHKAGSLAKKQNCFDDFISGAEYLVSAGYTQPSKLCIEGGSNGGLL--- 573
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDYGNPDKEEDFKYIYKISPIHNI-N 119
RPDL+ +A VGVMD+LRF+KFTIGHAW SDYG + EE+F ++ K SP+HN+
Sbjct 574 ----RPDLYGCALAHVGVMDMLRFHKFTIGHAWTSDYGCSENEEEFHWLIKYSPLHNVKR 629
Query 120 PEVVRSD---KQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYS---SPVSTPLIAHID 173
P ++D + P+ +++TADHDDRV P HSLK +A LQH + S SP P+I I+
Sbjct 630 PWEQQTDHLVQYPSTMLLTADHDDRVVPLHSLKLLATLQHVLCTSLDNSPQMNPIIGRIE 689
Query 174 TKAGHGGGKPLMKVLDEQALTYGFIASVLGLKWS 207
KAGHG G+P K++DE A Y F+A ++ W+
Sbjct 690 VKAGHGAGRPTQKMIDEAADRYSFMAKMVNASWT 723
> 7302143
Length=723
Score = 205 bits (522), Expect = 5e-53, Method: Compositional matrix adjust.
Identities = 94/158 (59%), Positives = 121/158 (76%), Gaps = 0/158 (0%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
N+RGGGEYG+ W+ +KQ ++DFQ+AAE+L Y+T D+LAI G SNGGLLVGA
Sbjct 564 NLRGGGEYGMEWHNGGRMLSKQNVFNDFQAAAEFLTKNNYTTKDRLAIQGASNGGLLVGA 623
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDYGNPDKEEDFKYIYKISPIHNINP 120
C NQRPDLF A VA+VGVMD+LRF+KFTIGHAW SDYGNPD++ F + K SP+HN++
Sbjct 624 CINQRPDLFGAAVAQVGVMDMLRFHKFTIGHAWCSDYGNPDEKVHFANLIKFSPLHNVHI 683
Query 121 EVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAV 158
+ + + P+ L++TADHDDRVSP HS K++A LQ AV
Sbjct 684 PLNPNQEYPSTLILTADHDDRVSPLHSYKFVAALQEAV 721
> At1g69020
Length=798
Score = 103 bits (257), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 67/208 (32%), Positives = 104/208 (50%), Gaps = 13/208 (6%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
++RGGG W+K+ + KQ S DF +A+YL KGY LA +G S G +L A
Sbjct 594 DVRGGGSGEFSWHKSGTRSLKQNSIQDFIYSAKYLVEKGYVHRHHLAAVGYSAGAILPAA 653
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVS-----DYGNPDKEEDFKYIYKISPI 115
N P LF+AV+ KV +D+L N + + ++ ++GNPD + DF I SP
Sbjct 654 AMNMHPSLFQAVILKVPFVDVL--NTLSDPNLPLTLLDHEEFGNPDNQTDFGSILSYSPY 711
Query 116 HNINPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYSSPVSTPLIAHIDTK 175
I +V P++LV T+ HD RV + K++A+++ + + S +I +
Sbjct 712 DKIRKDVC----YPSMLVTTSFHDSRVGVWEGAKWVAKIRDSTCHD--CSRAVILKTNMN 765
Query 176 AGHGGGKPLMKVLDEQALTYGFIASVLG 203
GH G +E A Y F+ V+G
Sbjct 766 GGHFGEGGRYAQCEETAFDYAFLLKVMG 793
> At1g50380
Length=710
Score = 99.8 bits (247), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 64/207 (30%), Positives = 103/207 (49%), Gaps = 12/207 (5%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
++RGGGE G WY+ K+ ++ DF + AE L Y + ++L + G S GGLL+GA
Sbjct 508 HVRGGGEMGRQWYENGKLLKKKNTFTDFIACAERLIELKYCSKEKLCMEGRSAGGLLMGA 567
Query 61 CANQRPDLFRAVVAKVGVMDLLRFN---KFTIGHAWVSDYGNPDKEEDFKYIYKISPIHN 117
N RPDLF+ V+A V +D+L + + ++G+P KEE + Y+ SP+ N
Sbjct 568 VVNMRPDLFKVVIAGVPFVDVLTTMLDPTIPLTTSEWEEWGDPRKEEFYFYMKSYSPVDN 627
Query 118 INPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYSSPVSTPLIAHIDTKAG 177
+ + P +LV +D RV K++A+L+ + L+ + AG
Sbjct 628 VTAQ-----NYPNMLVTAGLNDPRVMYSEPGKWVAKLREM----KTDNNVLLFKCELGAG 678
Query 178 HGGGKPLMKVLDEQALTYGFIASVLGL 204
H + L E A T+ F+ VL +
Sbjct 679 HFSKSGRFEKLQEDAFTFAFMMKVLDM 705
> Hs14727117
Length=638
Score = 70.5 bits (171), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 48/176 (27%), Positives = 81/176 (46%), Gaps = 18/176 (10%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
++RGGGE G+ W+ K D ++ + L +G+S P + S GG+L GA
Sbjct 419 HVRGGGELGLQWHADGRLTKKLNGLADLEACIKTLHGQGFSQPSLTTLTAFSAGGVLAGA 478
Query 61 CANQRPDLFRAVVAKVGVMDLLRF---NKFTIGHAWVSDYGNPDKEEDFK-YIYKISPIH 116
N P+L RAV + +D+L + + ++GNP +E K YI + P
Sbjct 479 LCNSNPELVRAVTLEAPFLDVLNTMMDTTLPLTLEELEEWGNPSSDEKHKNYIKRYCPYQ 538
Query 117 NINPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAV---------GYSSP 163
NI P+ P++ + ++D+RV + Y +L+ A+ GY +P
Sbjct 539 NIKPQ-----HYPSIHITAYENDERVPLKGIVSYTEKLKEAIAEHAKDTGEGYQTP 589
> At5g66960
Length=792
Score = 58.5 bits (140), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 52/158 (32%), Positives = 77/158 (48%), Gaps = 8/158 (5%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
++RGGG G W++ K S D+ A+YL ++LA G S GGL+V +
Sbjct 596 DVRGGGGKGKKWHQDGRGAKKLNSIKDYIQCAKYLVENNIVEENKLAGWGYSAGGLVVAS 655
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDY---GNPDKEEDFKYIYKISPIHN 117
N PDLF+A V KV +D + I DY G P DF I + SP N
Sbjct 656 AINHCPDLFQAAVLKVPFLDPTHTLIYPILPLTAEDYEEFGYPGDINDFHAIREYSPYDN 715
Query 118 INPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQ 155
I +V+ PAVLV T+ + R + + K++A+++
Sbjct 716 IPKDVL----YPAVLV-TSSFNTRFGVWEAAKWVARVR 748
> CE01231
Length=629
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 43/88 (48%), Gaps = 0/88 (0%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
N RG +G + + K DD + A+ L +G +++ I G S+GG L+ +
Sbjct 424 NYRGSTGFGTEFRRMLYKNCGVADRDDMLNGAKALVEQGKVDAEKVLITGSSSGGYLILS 483
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFT 88
C ++ +A V+ GV DLL ++ T
Sbjct 484 CLISPKNIIKAAVSVYGVADLLALDEDT 511
> At5g36210
Length=678
Score = 42.4 bits (98), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 56/206 (27%), Positives = 92/206 (44%), Gaps = 24/206 (11%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
N G YG + + +++ V DD A+YL + G + +L I GGS GG A
Sbjct 476 NYGGSTGYGREYRERLLRQWGIVDVDDCCGCAKYLVSSGKADVKRLCISGGSAGGYTTLA 535
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDYGNP--DKEEDFKYIYKISPIHNI 118
R D+F+A + GV DL + GH + S Y + E+DF Y+ SPI+ +
Sbjct 536 SLAFR-DVFKAGASLYGVADLKMLKE--EGHKFESRYIDNLVGDEKDF---YERSPINFV 589
Query 119 NPEVVRSDKQPAVLVMTADHDDR-VSPFHSLKYIAQLQHAVGYSSPVSTPLIAHIDTKAG 177
DK +++ +D+ V+P S K L+ PV+ L+ + + G
Sbjct 590 -------DKFSCPIILFQGLEDKVVTPDQSRKIYEALKKK---GLPVA--LVEYEGEQHG 637
Query 178 HGGGKPLMKVLDEQALTYGFIASVLG 203
+ + L++Q + F A V+G
Sbjct 638 FRKAENIKYTLEQQMV---FFARVVG 660
> Hs13640732
Length=732
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 38/126 (30%), Positives = 59/126 (46%), Gaps = 14/126 (11%)
Query 27 DFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLLRFNK 86
D Q A E + + + +A+MGGS+GG + Q P+ +RA VA+ V+++
Sbjct 562 DVQFAVEQVLQEEHFDASHVALMGGSHGGFISCHLIGQYPETYRACVARNPVINIASMLG 621
Query 87 FTIGHAW-VSDYGNP---DKEEDFKYIYKI---SPIHNINPEVVRSDKQPAVLVMTADHD 139
T W V + G P D D ++ SPI I P+V K P +L++ +
Sbjct 622 STDIPDWCVVEAGFPFSSDCLPDLSVWAEMLDKSPIRYI-PQV----KTPLLLML--GQE 674
Query 140 DRVSPF 145
DR PF
Sbjct 675 DRRVPF 680
> Hs9951917
Length=732
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 38/126 (30%), Positives = 59/126 (46%), Gaps = 14/126 (11%)
Query 27 DFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLLRFNK 86
D Q A E + + + +A+MGGS+GG + Q P+ +RA VA+ V+++
Sbjct 562 DVQFAVEQVLQEEHFDASHVALMGGSHGGFISCHLIGQYPETYRACVARNPVINIASMLG 621
Query 87 FTIGHAW-VSDYGNP---DKEEDFKYIYKI---SPIHNINPEVVRSDKQPAVLVMTADHD 139
T W V + G P D D ++ SPI I P+V K P +L++ +
Sbjct 622 STDIPDWCVVEAGFPFSSDCLPDLSVWAEMLDKSPIRYI-PQV----KTPLLLML--GQE 674
Query 140 DRVSPF 145
DR PF
Sbjct 675 DRRVPF 680
> At5g24260
Length=746
Score = 38.5 bits (88), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 40/157 (25%), Positives = 71/157 (45%), Gaps = 15/157 (9%)
Query 3 RGGGEYGVGWYKAAIKENK-QVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGAC 61
RG G+ +++ +K N V +D + A++L +G + PD + + G S GG L
Sbjct 566 RGTARRGLK-FESWMKHNCGYVDAEDQVTGAKWLIEQGLAKPDHIGVYGWSYGGYLSATL 624
Query 62 ANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDY-GNPDKEEDFKYIYKISPIHNINP 120
+ P++F V+ V ++ F + Y G P +EE + K S +H++
Sbjct 625 LTRYPEIFNCAVSGAPVTSWDGYDSF-----YTEKYMGLPTEEERY---LKSSVMHHVGN 676
Query 121 EVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHA 157
+DKQ +LV D+ V H+ + + L A
Sbjct 677 ---LTDKQKLMLVHGMI-DENVHFRHTARLVNALVEA 709
> CE19550
Length=745
Score = 36.6 bits (83), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 36/164 (21%), Positives = 66/164 (40%), Gaps = 16/164 (9%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKG-YSTPDQLAIMGGSNGGLLVG 59
N RG +G + +A + D +A + K + D++ + GGS+GG LV
Sbjct 548 NFRGSLGFGDDFIRALPGNCGDMDVKDVHNAVLTVLDKNPRISRDKVVLFGGSHGGFLVS 607
Query 60 ACANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWV--------SDYGNPDKEEDFKYIYK 111
Q P +++ VA V+++ + T W D+ E + ++
Sbjct 608 HLIGQYPGFYKSCVALNPVVNIATMHDITDIPEWCYFEGTGEYPDWTKITTTEQREKMFN 667
Query 112 ISPIHNINPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQ 155
SPI ++ + L++ + D RV P H +I L+
Sbjct 668 SSPIAHV------ENATTPYLLLIGEKDLRVVP-HYRAFIRALK 704
> CE00170
Length=761
Score = 34.7 bits (78), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 48/211 (22%), Positives = 81/211 (38%), Gaps = 23/211 (10%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
N RG +G A E + + D A E+ +KG + ++A+MGGS GG
Sbjct 486 NFRGSTGFGKRLTNAGNGEWGRKMHFDILDAVEFAVSKGIANRSEVAVMGGSYGGYETLV 545
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDYGNPDK--------EEDFKYIYKI 112
P F V VG +L+ + I W+ + K EE + +
Sbjct 546 ALTFTPQTFACGVDIVGPSNLISLVQ-AIPPYWLGFRKDLIKMVGADISDEEGRQSLQSR 604
Query 113 SPIHNINPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYSSPVSTPLIAHI 172
SP+ + R K ++++ +D RV S +++A L+ P+ +
Sbjct 605 SPLFFAD----RVTK--PIMIIQGANDPRVKQAESDQFVAALEKK-------HIPVTYLL 651
Query 173 DTKAGHGGGKPLMKVLDEQALTYGFIASVLG 203
GHG KP +++ F+ LG
Sbjct 652 YPDEGHGVRKP-QNSMEQHGHIETFLQQCLG 681
> Hs18450280
Length=882
Score = 33.9 bits (76), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 29/109 (26%), Positives = 48/109 (44%), Gaps = 7/109 (6%)
Query 3 RGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTK-GYSTPDQLAIMGGSNGGLLVGAC 61
RG G+ + A + Q+ DD +YL ++ + D++ I G S GG L
Sbjct 689 RGSCHRGLKFEGAFKYKMGQIEIDDQVEGLQYLASRYDFIDLDRVGIHGWSYGGYLSLMA 748
Query 62 ANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDY-GNPDKEEDFKYI 109
QR D+FR +A V + ++ + Y G+PD+ E Y+
Sbjct 749 LMQRSDIFRVAIAGAPVTLWIFYDT-----GYTERYMGHPDQNEQGYYL 792
> Hs22041021
Length=746
Score = 32.0 bits (71), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 42/165 (25%), Positives = 64/165 (38%), Gaps = 16/165 (9%)
Query 3 RGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGACA 62
RG G G+ + + V D +A ++L Y +L+I G GG +
Sbjct 552 RGSGFQGLKILQEIHRRLGSVEVKDQITAVKFLLKLPYIDSKRLSIFGKGYGGYIASMIL 611
Query 63 NQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDY-GNPDKEEDFKYIYK-ISPIHNINP 120
LF+ + DL + A+ Y G P KEE Y+ S +HN
Sbjct 612 KSDEKLFKCGSVVAPITDLKLY-----ASAFSERYLGMPSKEES---TYQAASVLHN--- 660
Query 121 EVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHA-VGYSSPV 164
V K+ +L++ D +V HS + I L A V Y+ V
Sbjct 661 --VHGLKEENILIIHGTADTKVHFQHSAELIKHLIKAGVNYTMQV 703
> At5g20520
Length=340
Score = 31.6 bits (70), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 39/155 (25%), Positives = 62/155 (40%), Gaps = 17/155 (10%)
Query 27 DFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAK---VGVMD--- 80
D Q+A ++L + ++ + G S GG + PD A++ + ++D
Sbjct 161 DAQAALDHLSGRTDIDTSRIVVFGRSLGGAVGAVLTKNNPDKVSALILENTFTSILDMAG 220
Query 81 -LLRFNKFTIGHAWVSDYGNPDKEEDFKYIYKISPIHNINPEVVRSDKQPAVLVMTADHD 139
LL F K+ IG G+ K SP I+ + KQP VL ++ D
Sbjct 221 VLLPFLKWFIG-------GSGTKSLKLLNFVVRSPWKTID--AIAEIKQP-VLFLSGLQD 270
Query 140 DRVSPFHSLKYIAQLQHAVGYSSPVSTPLIAHIDT 174
+ V PFH A+ + V P H+DT
Sbjct 271 EMVPPFHMKMLYAKAAARNPQCTFVEFPSGMHMDT 305
> Hs21264616
Length=715
Score = 30.0 bits (66), Expect = 3.4, Method: Composition-based stats.
Identities = 22/80 (27%), Positives = 34/80 (42%), Gaps = 1/80 (1%)
Query 89 IGHAWVSDYGNPDKEEDFKYIYKISPIHNINPEVVRSDKQPAVLVMTADHDDRVSPFHSL 148
+G A D G P ++ KY SP H + P +K + M +D+ + ++
Sbjct 546 LGEARPEDAGAPFEQAGQKYWGPASPTHKLPPS-FPGNKDELMQHMDEVNDELIRKISNI 604
Query 149 KYIAQLQHAVGYSSPVSTPL 168
+ Q V S PVS PL
Sbjct 605 RAQPQRHFRVERSQPVSQPL 624
> At2g01950
Length=1143
Score = 30.0 bits (66), Expect = 3.7, Method: Composition-based stats.
Identities = 14/52 (26%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 116 HNINPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYSSPVSTP 167
HN P ++ D + + +++ D + RVS F + I+ L + S+ TP
Sbjct 956 HNCIPHIIHRDMKSSNVLLDQDMEARVSDFGMARLISALDTHLSVSTLAGTP 1007
> CE05324
Length=829
Score = 28.9 bits (63), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 42/166 (25%), Positives = 66/166 (39%), Gaps = 29/166 (17%)
Query 1 NIRGGGEYGVGW-YKAAI--KENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLL 57
++RG G G GW K A+ K D ++ T G+ D++A+MG S GG L
Sbjct 639 DVRGTG--GRGWDVKEAVYRKLGDAEVVDTLDMIRAFINTFGFIDEDRIAVMGWSYGGFL 696
Query 58 VGACANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDYGNPDKEEDFKYIYKISPIHN 117
+K+ + D K I A V+D+ D +Y+ + P N
Sbjct 697 ---------------TSKIAIKDQGELVKCAISIAPVTDFKYYDSAYTERYLGQ--PAEN 739
Query 118 ----INPEVV---RSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQH 156
IN V+ R+ L+ + DD V +S ++ LQ
Sbjct 740 LQGYINTNVIPHARNVTNVKYLLAHGERDDNVHYQNSARWSEALQQ 785
> HsM12232453
Length=743
Score = 28.9 bits (63), Expect = 8.6, Method: Composition-based stats.
Identities = 21/80 (26%), Positives = 33/80 (41%), Gaps = 1/80 (1%)
Query 89 IGHAWVSDYGNPDKEEDFKYIYKISPIHNINPEVVRSDKQPAVLVMTADHDDRVSPFHSL 148
+G A D G P ++ KY SP H + P +K + M +D+ + ++
Sbjct 574 LGEARPEDAGAPFEQAGQKYWGPASPTHKLPPS-FPGNKDELMQRMDEVNDELIRKISNI 632
Query 149 KYIAQLQHAVGYSSPVSTPL 168
+ Q V S P S PL
Sbjct 633 RAQPQRHFRVERSQPASQPL 652
Lambda K H
0.317 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3969005466
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40