bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1112_orf1
Length=121
Score E
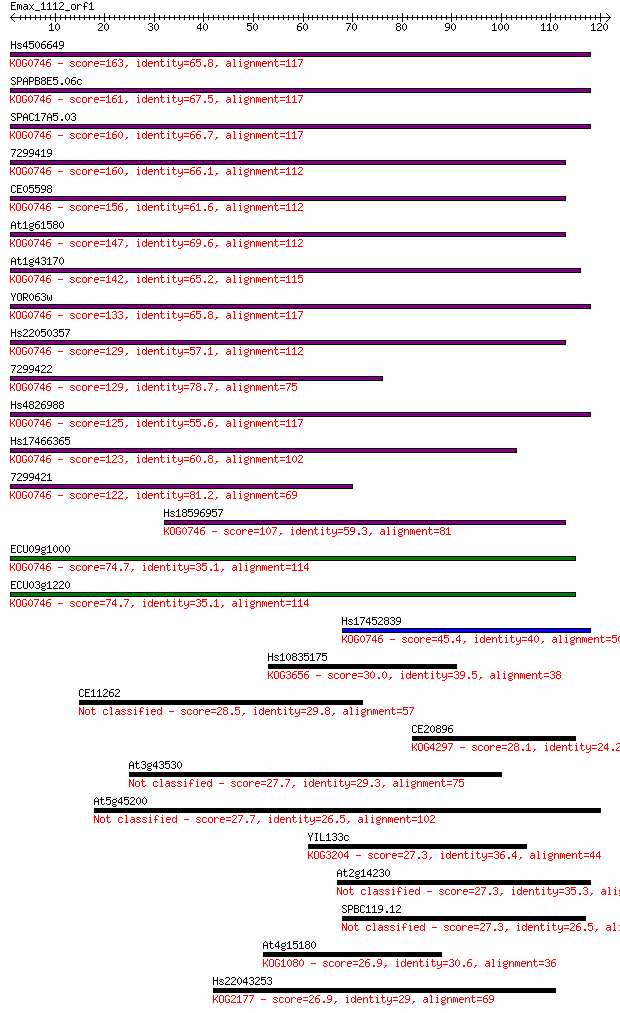
Sequences producing significant alignments: (Bits) Value
Hs4506649 163 7e-41
SPAPB8E5.06c 161 2e-40
SPAC17A5.03 160 4e-40
7299419 160 6e-40
CE05598 156 7e-39
At1g61580 147 5e-36
At1g43170 142 1e-34
YOR063w 133 9e-32
Hs22050357 129 1e-30
7299422 129 1e-30
Hs4826988 125 1e-29
Hs17466365 123 9e-29
7299421 122 2e-28
Hs18596957 107 6e-24
ECU09g1000 74.7 3e-14
ECU03g1220 74.7 3e-14
Hs17452839 45.4 2e-05
Hs10835175 30.0 0.96
CE11262 28.5 3.5
CE20896 28.1 3.7
At3g43530 27.7 5.7
At5g45200 27.7 6.1
YIL133c 27.3 7.4
At2g14230 27.3 7.6
SPBC119.12 27.3 7.8
At4g15180 26.9 8.2
Hs22043253 26.9 9.2
> Hs4506649
Length=403
Score = 163 bits (413), Expect = 7e-41, Method: Compositional matrix adjust.
Identities = 77/120 (64%), Positives = 99/120 (82%), Gaps = 3/120 (2%)
Query 1 MTHIVREVERPGSKLHKKEVVEAVTIVESPPMICVGVVGYIETPRGLRALSSVWAGHLSE 60
MTHIVREV+RPGSK++KKEVVEAVTIVE+PPM+ VG+VGY+ETPRGLR +V+A H+S+
Sbjct 53 MTHIVREVDRPGSKVNKKEVVEAVTIVETPPMVVVGIVGYVETPRGLRTFKTVFAEHISD 112
Query 61 ECRRRFYKNWYKSKKKAFTRYSRKYA-ENNKMQAEID--TIKQHCSVVRAICHTQPSKTP 117
EC+RRFYKNW+KSKKKAFT+Y +K+ E+ K Q E D ++K++C V+R I HTQ P
Sbjct 113 ECKRRFYKNWHKSKKKAFTKYCKKWQDEDGKKQLEKDFSSMKKYCQVIRVIAHTQMRLLP 172
> SPAPB8E5.06c
Length=388
Score = 161 bits (408), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 79/118 (66%), Positives = 106/118 (89%), Gaps = 1/118 (0%)
Query 1 MTHIVREVERPGSKLHKKEVVEAVTIVESPPMICVGVVGYIETPRGLRALSSVWAGHLSE 60
MTHIVR+++RPGSK+HK+E++EAVTI+E+PPM+ VGVVGY+ETPRGLR+L++VWA HLSE
Sbjct 53 MTHIVRDLDRPGSKMHKREILEAVTIIETPPMVVVGVVGYVETPRGLRSLTTVWAEHLSE 112
Query 61 ECRRRFYKNWYKSKKKAFTRYSRKYAENNK-MQAEIDTIKQHCSVVRAICHTQPSKTP 117
E +RRFYKNW+KSKKKAFT+Y++KYAE+ + + E++ IK++CSVVR + HTQ KTP
Sbjct 113 EVKRRFYKNWFKSKKKAFTKYAKKYAESTQSINRELERIKKYCSVVRVLAHTQIRKTP 170
> SPAC17A5.03
Length=388
Score = 160 bits (406), Expect = 4e-40, Method: Compositional matrix adjust.
Identities = 78/118 (66%), Positives = 106/118 (89%), Gaps = 1/118 (0%)
Query 1 MTHIVREVERPGSKLHKKEVVEAVTIVESPPMICVGVVGYIETPRGLRALSSVWAGHLSE 60
MTHIVR+++RPGSK+HK+E++EAVT++E+PPM+ VGVVGY+ETPRGLR+L++VWA HLSE
Sbjct 53 MTHIVRDLDRPGSKMHKREILEAVTVIETPPMVVVGVVGYVETPRGLRSLTTVWAEHLSE 112
Query 61 ECRRRFYKNWYKSKKKAFTRYSRKYAENNK-MQAEIDTIKQHCSVVRAICHTQPSKTP 117
E +RRFYKNW+KSKKKAFT+Y++KYAE+ + + E++ IK++CSVVR + HTQ KTP
Sbjct 113 EVKRRFYKNWFKSKKKAFTKYAKKYAESTQSINRELERIKKYCSVVRVLAHTQIRKTP 170
> 7299419
Length=403
Score = 160 bits (404), Expect = 6e-40, Method: Compositional matrix adjust.
Identities = 74/115 (64%), Positives = 94/115 (81%), Gaps = 3/115 (2%)
Query 1 MTHIVREVERPGSKLHKKEVVEAVTIVESPPMICVGVVGYIETPRGLRALSSVWAGHLSE 60
MTHIVRE +RPGSK++KKEVVEAVT++E+PPMI VG VGYIETP GLRAL +VWA HLSE
Sbjct 40 MTHIVREADRPGSKINKKEVVEAVTVLETPPMIVVGAVGYIETPFGLRALVNVWAQHLSE 99
Query 61 ECRRRFYKNWYKSKKKAFTRYSRKYAEN---NKMQAEIDTIKQHCSVVRAICHTQ 112
ECRRRFYKNWYKSKKKAFT+ S+K+ ++ ++ + + ++C V+R I H+Q
Sbjct 100 ECRRRFYKNWYKSKKKAFTKASKKWTDDLGKKSIENDFRKMLRYCKVIRVIAHSQ 154
> CE05598
Length=401
Score = 156 bits (395), Expect = 7e-39, Method: Compositional matrix adjust.
Identities = 69/115 (60%), Positives = 95/115 (82%), Gaps = 3/115 (2%)
Query 1 MTHIVREVERPGSKLHKKEVVEAVTIVESPPMICVGVVGYIETPRGLRALSSVWAGHLSE 60
MTHIVR+V++PGSK++KKEVVEAVTIVE+PPM+ GV GY++TP+G RAL+++WA HLSE
Sbjct 53 MTHIVRDVDKPGSKVNKKEVVEAVTIVETPPMVIAGVTGYVDTPQGPRALTTIWAEHLSE 112
Query 61 ECRRRFYKNWYKSKKKAFTRYSRKYAENNK---MQAEIDTIKQHCSVVRAICHTQ 112
E RRRFY NW KSKKKAFT+Y++K+ + + ++A+ +K++CS +R I HTQ
Sbjct 113 EARRRFYSNWAKSKKKAFTKYAKKWQDEDGKKLIEADFAKLKKYCSSIRVIAHTQ 167
> At1g61580
Length=390
Score = 147 bits (371), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 78/115 (67%), Positives = 99/115 (86%), Gaps = 3/115 (2%)
Query 1 MTHIVREVERPGSKLHKKEVVEAVTIVESPPMICVGVVGYIETPRGLRALSSVWAGHLSE 60
MTHIVR+VE+PGSKLHKKE EAVTI+E+PPM+ VGVVGY++TPRGLR+L +VWA HLSE
Sbjct 53 MTHIVRDVEKPGSKLHKKETCEAVTIIETPPMVVVGVVGYVKTPRGLRSLCTVWAQHLSE 112
Query 61 ECRRRFYKNWYKSKKKAFTRYSRKY-AENNK--MQAEIDTIKQHCSVVRAICHTQ 112
E RRRFYKNW KSKKKAFTRYS+K+ E K +Q++++ +K++CSV+R + HTQ
Sbjct 113 ELRRRFYKNWAKSKKKAFTRYSKKHETEEGKKDIQSQLEKMKKYCSVIRVLAHTQ 167
> At1g43170
Length=389
Score = 142 bits (359), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 75/118 (63%), Positives = 99/118 (83%), Gaps = 3/118 (2%)
Query 1 MTHIVREVERPGSKLHKKEVVEAVTIVESPPMICVGVVGYIETPRGLRALSSVWAGHLSE 60
MTHIVREVE+PGSKLHKKE EAVTI+E+P M+ VGVV Y++TPRGLR+L++VWA HLSE
Sbjct 53 MTHIVREVEKPGSKLHKKETCEAVTIIETPAMVVVGVVAYVKTPRGLRSLNTVWAQHLSE 112
Query 61 ECRRRFYKNWYKSKKKAFTRYSRKY-AENNK--MQAEIDTIKQHCSVVRAICHTQPSK 115
E RRRFYKNW KSKKKAFT Y+++Y +E+ K +QA+++ +K++ +V+R + HTQ K
Sbjct 113 EVRRRFYKNWAKSKKKAFTGYAKQYDSEDGKKGIQAQLEKMKKYATVIRVLAHTQIRK 170
> YOR063w
Length=387
Score = 133 bits (334), Expect = 9e-32, Method: Compositional matrix adjust.
Identities = 77/118 (65%), Positives = 101/118 (85%), Gaps = 1/118 (0%)
Query 1 MTHIVREVERPGSKLHKKEVVEAVTIVESPPMICVGVVGYIETPRGLRALSSVWAGHLSE 60
MT IVR+++RPGSK HK+EVVEAVT+V++PP++ VGVVGY+ETPRGLR+L++VWA HLS+
Sbjct 53 MTTIVRDLDRPGSKFHKREVVEAVTVVDTPPVVVVGVVGYVETPRGLRSLTTVWAEHLSD 112
Query 61 ECRRRFYKNWYKSKKKAFTRYSRKYAENNK-MQAEIDTIKQHCSVVRAICHTQPSKTP 117
E +RRFYKNWYKSKKKAFT+YS KYA++ ++ E+ IK++ SVVR + HTQ KTP
Sbjct 113 EVKRRFYKNWYKSKKKAFTKYSAKYAQDGAGIERELARIKKYASVVRVLVHTQIRKTP 170
> Hs22050357
Length=432
Score = 129 bits (325), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 64/115 (55%), Positives = 91/115 (79%), Gaps = 3/115 (2%)
Query 1 MTHIVREVERPGSKLHKKEVVEAVTIVESPPMICVGVVGYIETPRGLRALSSVWAGHLSE 60
MT+I+REV+RPGSK++KKEV+EAVTIVE PPM+ V +VG++ETPRGL+ ++V+A H+S+
Sbjct 88 MTNIMREVDRPGSKVNKKEVIEAVTIVEIPPMVVVAIVGFVETPRGLQTFNTVFAEHISD 147
Query 61 ECRRRFYKNWYKSKKKAFTRYSRK-YAENNKMQAEID--TIKQHCSVVRAICHTQ 112
C+R FYKN +KSKKKAFT+Y +K E+ K Q E D ++K++ V+ I +TQ
Sbjct 148 ACKRHFYKNRHKSKKKAFTKYCKKCQDEDGKKQLEKDFSSMKKYRQVICIIAYTQ 202
> 7299422
Length=148
Score = 129 bits (324), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 59/75 (78%), Positives = 67/75 (89%), Gaps = 0/75 (0%)
Query 1 MTHIVREVERPGSKLHKKEVVEAVTIVESPPMICVGVVGYIETPRGLRALSSVWAGHLSE 60
MTHIVRE +RPGSK++KKEVVEAVT++E+PPMI VG VGYIETP GLRAL +VWA HLSE
Sbjct 40 MTHIVREADRPGSKINKKEVVEAVTVLETPPMIVVGAVGYIETPFGLRALVNVWAQHLSE 99
Query 61 ECRRRFYKNWYKSKK 75
ECRRRFYKNWY S+
Sbjct 100 ECRRRFYKNWYVSED 114
> Hs4826988
Length=407
Score = 125 bits (315), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 65/120 (54%), Positives = 92/120 (76%), Gaps = 3/120 (2%)
Query 1 MTHIVREVERPGSKLHKKEVVEAVTIVESPPMICVGVVGYIETPRGLRALSSVWAGHLSE 60
MTH +REV RPG K+ K+E VEAVTIVE+PP++ VGVVGY+ TPRGLR+ +++A HLS+
Sbjct 53 MTHTLREVHRPGLKISKREEVEAVTIVETPPLVVVGVVGYVATPRGLRSFKTIFAEHLSD 112
Query 61 ECRRRFYKNWYKSKKKAFTRYSRKYAENN---KMQAEIDTIKQHCSVVRAICHTQPSKTP 117
ECRRRFYK+W+KSKKKAFT+ +++ + + ++Q + +K++C V+R I HTQ P
Sbjct 113 ECRRRFYKDWHKSKKKAFTKACKRWRDTDGKKQLQKDFAAMKKYCKVIRVIVHTQMKLLP 172
> Hs17466365
Length=251
Score = 123 bits (308), Expect = 9e-29, Method: Compositional matrix adjust.
Identities = 62/105 (59%), Positives = 82/105 (78%), Gaps = 4/105 (3%)
Query 1 MTHIVREVERPGSKLHKKEVVEAVTIVESPPMICVGVVGYIETPRGLRALSSVWAGHLSE 60
MTHIV EV+RPGSK++KKEVVEAVTIVE M+ G+VGY+ TPRGL +V+ H+S+
Sbjct 1 MTHIVWEVDRPGSKVNKKEVVEAVTIVE-ITMVVAGIVGYVGTPRGLWTFKTVFTEHISD 59
Query 61 ECRRRFYKNWYKSKKKAFTRYSRKY-AENNKMQAEID--TIKQHC 102
EC+R FYKNW+KSKKKAFT+Y +K+ E+ K Q E D ++K++C
Sbjct 60 ECKRHFYKNWHKSKKKAFTKYCKKWLDEDGKKQLEKDFSSMKKYC 104
> 7299421
Length=125
Score = 122 bits (305), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 56/69 (81%), Positives = 63/69 (91%), Gaps = 0/69 (0%)
Query 1 MTHIVREVERPGSKLHKKEVVEAVTIVESPPMICVGVVGYIETPRGLRALSSVWAGHLSE 60
MTHIVRE +RPGSK++KKEVVEAVT++E+PPMI VG VGYIETP GLRAL +VWA HLSE
Sbjct 40 MTHIVREADRPGSKINKKEVVEAVTVLETPPMIVVGAVGYIETPFGLRALVNVWAQHLSE 99
Query 61 ECRRRFYKN 69
ECRRRFYKN
Sbjct 100 ECRRRFYKN 108
> Hs18596957
Length=133
Score = 107 bits (267), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 48/84 (57%), Positives = 66/84 (78%), Gaps = 3/84 (3%)
Query 32 MICVGVVGYIETPRGLRALSSVWAGHLSEECRRRFYKNWYKSKKKAFTRYSRKYA-ENNK 90
M+ VG+VGY+ETP+GLR +V+A H+S+EC+RRFYKNW+KSKKKAFT+Y +K+ EN K
Sbjct 1 MMVVGIVGYVETPQGLRTFKTVFAEHISDECKRRFYKNWHKSKKKAFTKYRKKWQDENGK 60
Query 91 MQAEID--TIKQHCSVVRAICHTQ 112
Q E D ++K++C V+ I HTQ
Sbjct 61 KQLEKDFSSMKKYCQVIHVIAHTQ 84
> ECU09g1000
Length=383
Score = 74.7 bits (182), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 40/115 (34%), Positives = 70/115 (60%), Gaps = 6/115 (5%)
Query 1 MTHIVR-EVERPGSKLHKKEVVEAVTIVESPPMICVGVVGYIETPRGLRALSSVWAGHLS 59
MTH+VR + + +K +E+++AVT++E+PPM+ G+VGY +T GLR L V A ++S
Sbjct 53 MTHVVRTKTQVAKNKQLSREIMDAVTVLEAPPMVVYGIVGYEKTVTGLRRLPIVTAAYVS 112
Query 60 EECRRRFYKNWYKSKKKAFTRYSRKYAENNKMQAEIDTIKQHCSVVRAICHTQPS 114
+ RR + N Y SK+ A ++ + + ++ ++ IK+ VR + TQP+
Sbjct 113 DGVLRRMFGNRYASKESA-----GQFCKGSVCESRVEMIKERAHCVRVLVQTQPT 162
> ECU03g1220
Length=383
Score = 74.7 bits (182), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 40/115 (34%), Positives = 70/115 (60%), Gaps = 6/115 (5%)
Query 1 MTHIVR-EVERPGSKLHKKEVVEAVTIVESPPMICVGVVGYIETPRGLRALSSVWAGHLS 59
MTH+VR + + +K +E+++AVT++E+PPM+ G+VGY +T GLR L V A ++S
Sbjct 53 MTHVVRTKTQVAKNKQLSREIMDAVTVLEAPPMVVYGIVGYEKTVTGLRRLPIVTAAYVS 112
Query 60 EECRRRFYKNWYKSKKKAFTRYSRKYAENNKMQAEIDTIKQHCSVVRAICHTQPS 114
+ RR + N Y SK+ A ++ + + ++ ++ IK+ VR + TQP+
Sbjct 113 DGVLRRMFGNRYASKESA-----GQFCKGSVCESRVEMIKERAHCVRVLVQTQPT 162
> Hs17452839
Length=180
Score = 45.4 bits (106), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 35/53 (66%), Gaps = 3/53 (5%)
Query 68 KNWYKSKKKAFTRYSRKYAENN---KMQAEIDTIKQHCSVVRAICHTQPSKTP 117
KNW+KS+KKAFT Y +K+ + + +++ +++K++C V+ I HTQ P
Sbjct 66 KNWHKSEKKAFTEYCKKWQDEDGEKQLEKNFNSMKKYCQVICIIAHTQMCLLP 118
> Hs10835175
Length=471
Score = 30.0 bits (66), Expect = 0.96, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 22/39 (56%), Gaps = 1/39 (2%)
Query 53 VWAGHLSEECRRRFYKNWYKSKKKAFTRYSR-KYAENNK 90
VW G+LS Y + K+ + AF+RY + +Y EN K
Sbjct 366 VWIGYLSSAVNPLVYTLFNKTYRSAFSRYIQCQYKENKK 404
> CE11262
Length=299
Score = 28.5 bits (62), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 17/61 (27%), Positives = 27/61 (44%), Gaps = 4/61 (6%)
Query 15 LHKKEVVEAVTI----VESPPMICVGVVGYIETPRGLRALSSVWAGHLSEECRRRFYKNW 70
LHK E+ I V + P + GY++ G+R VW S++ + KNW
Sbjct 183 LHKIEMFSGNAIMAENVFTDPNGYFDIAGYVDQSSGVRGRFWVWHECFSQKYHKDPCKNW 242
Query 71 Y 71
+
Sbjct 243 F 243
> CE20896
Length=458
Score = 28.1 bits (61), Expect = 3.7, Method: Composition-based stats.
Identities = 8/33 (24%), Positives = 18/33 (54%), Gaps = 0/33 (0%)
Query 82 SRKYAENNKMQAEIDTIKQHCSVVRAICHTQPS 114
S+++ N + + ++QH ++ A+C PS
Sbjct 276 SKRHPNNADISKTVSLVRQHHGIIHAVCSVAPS 308
> At3g43530
Length=615
Score = 27.7 bits (60), Expect = 5.7, Method: Composition-based stats.
Identities = 22/79 (27%), Positives = 37/79 (46%), Gaps = 5/79 (6%)
Query 25 TIVESPPMICVGVVGYIETPRGLRALSSVWAGHLSEECRRRFYKNWYKSKKKAFTRYSRK 84
T+V++P V G +T L + S + E + +N +KS K T +S+K
Sbjct 331 TVVQAPRKAAVEKKGKGKTAAALTSPSDEGLTEVVNE-MKNLMENGFKSMNKRMTNFSKK 389
Query 85 YAENNK----MQAEIDTIK 99
Y E +K M+ I +I+
Sbjct 390 YEEQDKRLKLMETAIKSIQ 408
> At5g45200
Length=1261
Score = 27.7 bits (60), Expect = 6.1, Method: Composition-based stats.
Identities = 27/104 (25%), Positives = 46/104 (44%), Gaps = 12/104 (11%)
Query 18 KEVVEAVTIVESPPMICVGVVGYIETPRGLRALSSVWAGHLSEECRRRF--YKNWYKSKK 75
KE VEA T+V P V V + +R L+ + L R Y+ W ++ +
Sbjct 96 KECVEAGTLVVFPVFYKVDV-------KIVRFLTGSFGEKLETLVLRHSERYEPWKQALE 148
Query 76 KAFTRYSRKYAENNKMQAEIDTIKQHCSVVRAICHTQPSKTPTG 119
++ ++ EN+ AE++ I +H V+ I T + P G
Sbjct 149 FVTSKTGKRVEENSDEGAEVEQIVEH---VKEILRTISGEIPRG 189
> YIL133c
Length=199
Score = 27.3 bits (59), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 23/44 (52%), Gaps = 3/44 (6%)
Query 61 ECRRRFYKNWYKSKKKAFTRYSRKYAENNKMQAEIDTIKQHCSV 104
E +R+ Y +KK+AFT +K A N AE D KQ ++
Sbjct 157 EAKRKVSSAEYYAKKRAFT---KKVASANATAAESDVAKQLAAL 197
> At2g14230
Length=365
Score = 27.3 bits (59), Expect = 7.6, Method: Composition-based stats.
Identities = 18/63 (28%), Positives = 24/63 (38%), Gaps = 12/63 (19%)
Query 67 YKNWYKSKKKAFTRYSRKYAENNKMQAEIDT-----IKQHCS-------VVRAICHTQPS 114
+ NW ++ K R+ +A NN A I +HCS VR C
Sbjct 114 HANWSQTSKATIDRWYETFASNNNRNARYHDPDGKGIYKHCSGQTSYKARVRKRCEKTGE 173
Query 115 KTP 117
KTP
Sbjct 174 KTP 176
> SPBC119.12
Length=401
Score = 27.3 bits (59), Expect = 7.8, Method: Composition-based stats.
Identities = 13/49 (26%), Positives = 23/49 (46%), Gaps = 0/49 (0%)
Query 68 KNWYKSKKKAFTRYSRKYAENNKMQAEIDTIKQHCSVVRAICHTQPSKT 116
+NW K+ +++ K E +Q E+D ++ S V + H KT
Sbjct 239 RNWQKAMDDVTEKFASKSKEYEDLQNELDATQKRLSRVSDLEHEVKEKT 287
> At4g15180
Length=2351
Score = 26.9 bits (58), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 11/36 (30%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 52 SVWAGHLSEECRRRFYKNWYKSKKKAFTRYSRKYAE 87
S+ + H S+ R Y++ Y SK + +Y RK+ +
Sbjct 337 SLHSDHYSQHSAERLYRDSYPSKNSSLEKYPRKHQD 372
> Hs22043253
Length=412
Score = 26.9 bits (58), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 28/69 (40%), Gaps = 4/69 (5%)
Query 42 ETPRGLRALSSVWAGHLSEECRRRFYKNWYKSKKKAFTRYSRKYAENNKMQAEIDTIKQH 101
E PRG SS A H S+E R+F N + + ++ K Q E K+H
Sbjct 69 EVPRG----SSAGAPHPSDEAERKFISNPQLGSLTEIAKQLQIRSKKRKRQEEKHVCKKH 124
Query 102 CSVVRAICH 110
V+ C
Sbjct 125 NQVLTFFCQ 133
Lambda K H
0.319 0.131 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1198419392
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40