bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1101_orf1
Length=141
Score E
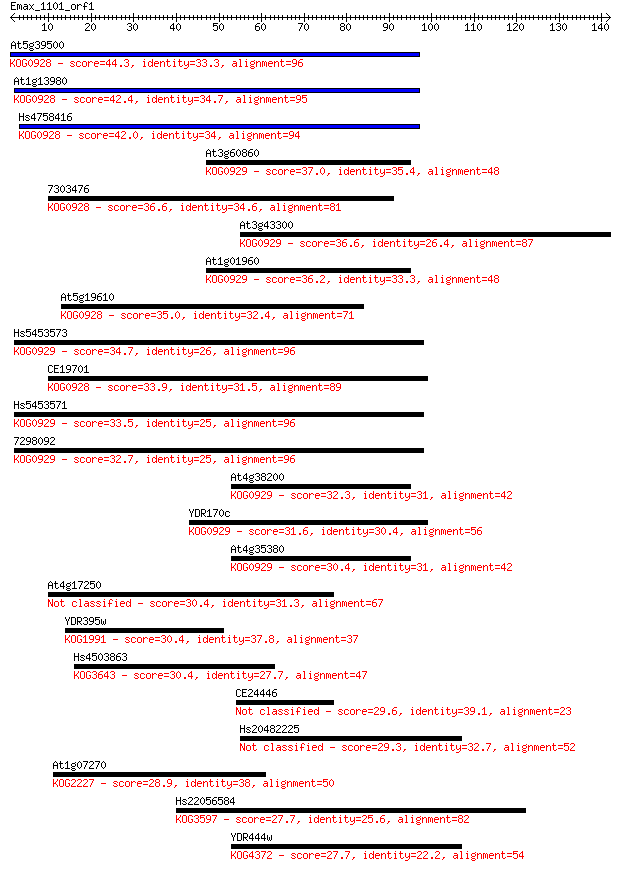
Sequences producing significant alignments: (Bits) Value
At5g39500 44.3 8e-05
At1g13980 42.4 3e-04
Hs4758416 42.0 4e-04
At3g60860 37.0 0.012
7303476 36.6 0.015
At3g43300 36.6 0.015
At1g01960 36.2 0.018
At5g19610 35.0 0.040
Hs5453573 34.7 0.064
CE19701 33.9 0.097
Hs5453571 33.5 0.13
7298092 32.7 0.24
At4g38200 32.3 0.31
YDR170c 31.6 0.47
At4g35380 30.4 1.1
At4g17250 30.4 1.2
YDR395w 30.4 1.2
Hs4503863 30.4 1.3
CE24446 29.6 1.9
Hs20482225 29.3 2.6
At1g07270 28.9 3.2
Hs22056584 27.7 6.7
YDR444w 27.7 7.2
> At5g39500
Length=1443
Score = 44.3 bits (103), Expect = 8e-05, Method: Composition-based stats.
Identities = 32/97 (32%), Positives = 50/97 (51%), Gaps = 6/97 (6%)
Query 1 SLTLRVVGDFIVFLRPYIKDEIERCL-YQFLRLATTNIGGPPKTLAAEAQEVALDMLREL 59
S +V + + LR +K ++E Y LR+A + G + + QEVA++ L +L
Sbjct 400 STVCSIVLNLYLNLRTELKVQLEAFFSYVLLRIAQSKHGS-----SYQQQEVAMEALVDL 454
Query 60 CSDASFGFELVLNYDSDIRRSNVFGVLVTLLLQLAAP 96
C +F E+ N+D DI SNVF + LL + A P
Sbjct 455 CRQHTFIAEVFANFDCDITCSNVFEDVSNLLSKNAFP 491
> At1g13980
Length=1451
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 33/100 (33%), Positives = 47/100 (47%), Gaps = 10/100 (10%)
Query 2 LTLRVVGDFIVFLRPYIKDEIERCLYQF-----LRLATTNIGGPPKTLAAEAQEVALDML 56
L L +V ++ L +++ E++ L F LRLA G + + QEVA++ L
Sbjct 396 LILSMVCSIVLNLYQHLRTELKLQLEAFFSCVILRLAQGKYGP-----SYQQQEVAMEAL 450
Query 57 RELCSDASFGFELVLNYDSDIRRSNVFGVLVTLLLQLAAP 96
C SF E+ N D DI SNVF L LL + P
Sbjct 451 VNFCRQKSFMVEMYANLDCDITCSNVFEELSNLLSKSTFP 490
> Hs4758416
Length=1859
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 53/96 (55%), Gaps = 8/96 (8%)
Query 3 TLRVVGDFIVF--LRPYIKDEIERCLYQFLRLATTNIGGPPKTLAAEAQEVALDMLRELC 60
+LRV F++F +R ++K ++E + + + + T PK + E +E+AL+ + +L
Sbjct 476 SLRVC--FLLFESMREHLKFQMEMYIKKLMEIITVE---NPK-MPYEMKEMALEAIVQLW 529
Query 61 SDASFGFELVLNYDSDIRRSNVFGVLVTLLLQLAAP 96
SF EL +NYD D SN+F L LL + A P
Sbjct 530 RIPSFVTELYINYDCDYYCSNLFEELTKLLSKNAFP 565
> At3g60860
Length=1793
Score = 37.0 bits (84), Expect = 0.012, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 28/48 (58%), Gaps = 0/48 (0%)
Query 47 EAQEVALDMLRELCSDASFGFELVLNYDSDIRRSNVFGVLVTLLLQLA 94
+ + + L L +LC D+ ++ LNYD D+ SN+F +V LL+ A
Sbjct 469 QQKMIVLRFLDKLCLDSQILVDIFLNYDCDVNSSNIFERMVNGLLKTA 516
> 7303476
Length=2040
Score = 36.6 bits (83), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 28/83 (33%), Positives = 46/83 (55%), Gaps = 6/83 (7%)
Query 10 FIVF--LRPYIKDEIERCLYQFLRLATTNIGGPPKTLAAEAQEVALDMLRELCSDASFGF 67
F++F LR ++K ++E L + + ++ PKT E +E+ALD L +L F
Sbjct 447 FLLFESLRGHLKFQLEAYLRKLSEIIASD---NPKT-PYEMRELALDNLLQLWRIPGFVT 502
Query 68 ELVLNYDSDIRRSNVFGVLVTLL 90
EL +NYD D+ +++F L LL
Sbjct 503 ELYINYDCDLYCTDMFESLTNLL 525
> At3g43300
Length=1669
Score = 36.6 bits (83), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 23/99 (23%), Positives = 43/99 (43%), Gaps = 12/99 (12%)
Query 55 MLRELCSDASFGFELVLNYDSDIRRSNVFGVLVTLLLQLA------------APRVPSLR 102
ML ++C D ++ +NYD D+ N+F +VT L ++A A + S++
Sbjct 467 MLEKVCKDPQMLVDVYVNYDCDLEAPNLFERMVTTLSKIAQGSQSADPNPAMASQTASVK 526
Query 103 GAYRFAEYDNEGGGGLMGASGTPLSHATLGQVATGPESS 141
G+ AE ++G P+ + V + E +
Sbjct 527 GSSLQAENSTRNANEDSASTGEPIETKSREDVPSNFEKA 565
> At1g01960
Length=1750
Score = 36.2 bits (82), Expect = 0.018, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 28/48 (58%), Gaps = 0/48 (0%)
Query 47 EAQEVALDMLRELCSDASFGFELVLNYDSDIRRSNVFGVLVTLLLQLA 94
+ + + L L +LC D+ ++ +NYD D+ SN+F +V LL+ A
Sbjct 457 QQKMIVLRFLDKLCVDSQILVDIFINYDCDVNSSNIFERMVNGLLKTA 504
> At5g19610
Length=1375
Score = 35.0 bits (79), Expect = 0.040, Method: Composition-based stats.
Identities = 23/71 (32%), Positives = 34/71 (47%), Gaps = 6/71 (8%)
Query 13 FLRPYIKDEIERCLYQFLRLATTNIGGPPKTLAAEAQEVALDMLRELCSDASFGFELVLN 72
FLR +++ ++E + F+ L T G QEVAL+ L C +F E +N
Sbjct 336 FLRKFMRLQLE-AFFSFVLLRVTAFTG-----FLPLQEVALEGLINFCRQPAFIVEAYVN 389
Query 73 YDSDIRRSNVF 83
YD D N+F
Sbjct 390 YDCDPMCRNIF 400
> Hs5453573
Length=1785
Score = 34.7 bits (78), Expect = 0.064, Method: Composition-based stats.
Identities = 25/97 (25%), Positives = 49/97 (50%), Gaps = 7/97 (7%)
Query 2 LTLRVVGDFIVFLRPYIKDEIERCLYQ-FLRLATTNIGGPPKTLAAEAQEVALDMLRELC 60
L+L + + + ++K +IE + FL + T+ T + E + + + L +C
Sbjct 454 LSLAIFLTLLSNFKMHLKMQIEVFFKEIFLNILETS------TSSFEHRWMVIQTLTRIC 507
Query 61 SDASFGFELVLNYDSDIRRSNVFGVLVTLLLQLAAPR 97
+DA ++ +NYD D+ +N+F LV L ++A R
Sbjct 508 ADAQCVVDIYVNYDCDLNAANIFERLVNDLSKIAQGR 544
> CE19701
Length=1820
Score = 33.9 bits (76), Expect = 0.097, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 48/97 (49%), Gaps = 13/97 (13%)
Query 10 FIVF--LRPYIKDEIERCLYQFLRLATT------NIGGPPKTLAAEAQEVALDMLRELCS 61
F++F +R ++K ++E L + + T N GG E +E+AL+ L +L
Sbjct 468 FLLFESMRMHMKFQLESYLKKLQSIVLTEEKQHENGGG-----GTEQKEMALESLVQLWR 522
Query 62 DASFGFELVLNYDSDIRRSNVFGVLVTLLLQLAAPRV 98
E+ LN+D D+ N+F L LL++ + P V
Sbjct 523 IPGLVTEMYLNFDCDLYCGNIFEDLTKLLVENSFPTV 559
> Hs5453571
Length=1849
Score = 33.5 bits (75), Expect = 0.13, Method: Composition-based stats.
Identities = 24/97 (24%), Positives = 49/97 (50%), Gaps = 7/97 (7%)
Query 2 LTLRVVGDFIVFLRPYIKDEIERCLYQ-FLRLATTNIGGPPKTLAAEAQEVALDMLRELC 60
L+L + + + ++K +IE + FL + T+ T + + + + + L +C
Sbjct 503 LSLSIFLTLLSNFKTHLKMQIEVFFKEIFLYILETS------TSSFDHKWMVIQTLTRIC 556
Query 61 SDASFGFELVLNYDSDIRRSNVFGVLVTLLLQLAAPR 97
+DA ++ +NYD D+ +N+F LV L ++A R
Sbjct 557 ADAQSVVDIYVNYDCDLNAANIFERLVNDLSKIAQGR 593
> 7298092
Length=1643
Score = 32.7 bits (73), Expect = 0.24, Method: Composition-based stats.
Identities = 24/97 (24%), Positives = 47/97 (48%), Gaps = 7/97 (7%)
Query 2 LTLRVVGDFIVFLRPYIKDEIERCLYQ-FLRLATTNIGGPPKTLAAEAQEVALDMLRELC 60
L+L + + + ++K +IE + FL + N + + E + + + L +C
Sbjct 386 LSLSIFVALLSNFKVHLKRQIEVFFKEIFLNILEAN------SSSFEHKWMVIQALTRIC 439
Query 61 SDASFGFELVLNYDSDIRRSNVFGVLVTLLLQLAAPR 97
+DA ++ +NYD D +N+F LV L ++A R
Sbjct 440 ADAQSVVDIYVNYDCDFSAANLFERLVNDLSKIAQGR 476
> At4g38200
Length=1698
Score = 32.3 bits (72), Expect = 0.31, Method: Composition-based stats.
Identities = 13/42 (30%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 53 LDMLRELCSDASFGFELVLNYDSDIRRSNVFGVLVTLLLQLA 94
L +L +C D + ++ +N+D D+ N+F +V LL+ A
Sbjct 412 LSLLENICHDPNLIIDIFVNFDCDVESPNIFERIVNGLLKTA 453
> YDR170c
Length=2009
Score = 31.6 bits (70), Expect = 0.47, Method: Composition-based stats.
Identities = 17/56 (30%), Positives = 28/56 (50%), Gaps = 0/56 (0%)
Query 43 TLAAEAQEVALDMLRELCSDASFGFELVLNYDSDIRRSNVFGVLVTLLLQLAAPRV 98
T ++ + L +++ +C+D E LNYD + NV + V L +LA RV
Sbjct 624 TSTSQQKRYFLSVIQRICNDPRTLVEFYLNYDCNPGMPNVMEITVDYLTRLALTRV 679
> At4g35380
Length=1711
Score = 30.4 bits (67), Expect = 1.1, Method: Composition-based stats.
Identities = 13/42 (30%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 53 LDMLRELCSDASFGFELVLNYDSDIRRSNVFGVLVTLLLQLA 94
L++L ++ D ++ +NYD D+ SN+ +V LL+ A
Sbjct 422 LNLLDKMSQDPQLMVDIFVNYDCDVESSNILERIVNGLLKTA 463
> At4g17250
Length=633
Score = 30.4 bits (67), Expect = 1.2, Method: Composition-based stats.
Identities = 21/71 (29%), Positives = 30/71 (42%), Gaps = 4/71 (5%)
Query 10 FIVFLRPYIKDEIERCLYQFL----RLATTNIGGPPKTLAAEAQEVALDMLRELCSDASF 65
F+ F+ Y I C + F +A GP T +E L L E + S
Sbjct 371 FLRFITVYCSSRILLCCFHFTFKSDSVANAESSGPLDTKGSEEPTCNLIELLEATTFISK 430
Query 66 GFELVLNYDSD 76
GFE + ++DSD
Sbjct 431 GFEGIRDFDSD 441
> YDR395w
Length=944
Score = 30.4 bits (67), Expect = 1.2, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 14 LRPYIKDEIERCLYQFLRLATTNIGGPPKTLAAEAQE 50
L P+ KD + QFLR+A + P +T +A QE
Sbjct 569 LSPFAKDLASNLVEQFLRIAQALVENPSETYSASDQE 605
> Hs4503863
Length=474
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 13/47 (27%), Positives = 24/47 (51%), Gaps = 0/47 (0%)
Query 16 PYIKDEIERCLYQFLRLATTNIGGPPKTLAAEAQEVALDMLRELCSD 62
PY+K+ ++R L + + GGPP + ++DM+ E+ D
Sbjct 35 PYVKETVDRLLKGYDIRLRPDFGGPPVDVGMRIDVASIDMVSEVNMD 81
> CE24446
Length=564
Score = 29.6 bits (65), Expect = 1.9, Method: Composition-based stats.
Identities = 9/23 (39%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 54 DMLRELCSDASFGFELVLNYDSD 76
+++RE+C+D ++ FE +N D D
Sbjct 222 ELIREMCADQAYSFEKTINEDMD 244
> Hs20482225
Length=316
Score = 29.3 bits (64), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 17/53 (32%), Positives = 31/53 (58%), Gaps = 1/53 (1%)
Query 55 MLRELCSDASFGFELVLNYDSDIRRSNVFGVLVT-LLLQLAAPRVPSLRGAYR 106
+L+ CSD F +++ + S I S V VL++ L + +A ++PSL G ++
Sbjct 190 LLKLSCSDTHFNGIVIMAFSSFIVISCVMIVLISYLCIFIAVLKMPSLEGRHK 242
> At1g07270
Length=507
Score = 28.9 bits (63), Expect = 3.2, Method: Composition-based stats.
Identities = 19/53 (35%), Positives = 29/53 (54%), Gaps = 4/53 (7%)
Query 11 IVFLRPYIKDEIERCLYQFLRLATTNIGGPPKTLAAEAQEVAL---DMLRELC 60
++ R Y KD+I R L + LR+ + + PK L A++VA DM + LC
Sbjct 301 VITFRAYSKDQILRILQERLRVLSY-VAFQPKALELCARKVAAASGDMRKALC 352
> Hs22056584
Length=1330
Score = 27.7 bits (60), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 37/82 (45%), Gaps = 8/82 (9%)
Query 40 PPKTLAAEAQEVALDMLRELCSDASFGFELVLNYDSDIRRSNVFGVLVTLLLQLAAPRVP 99
PP TL A + A+ + R + G + + + + +N G V++L ++
Sbjct 581 PPATLKVMAIQPAIQIHRSTGLHLAQGSAMPILPANLLVETNAVGQDVSVLFRVT----- 635
Query 100 SLRGAYRFAEYDNEGGGGLMGA 121
GA +F E +G GG+ GA
Sbjct 636 ---GALKFGELKKQGAGGVEGA 654
> YDR444w
Length=687
Score = 27.7 bits (60), Expect = 7.2, Method: Composition-based stats.
Identities = 12/54 (22%), Positives = 29/54 (53%), Gaps = 0/54 (0%)
Query 53 LDMLRELCSDASFGFELVLNYDSDIRRSNVFGVLVTLLLQLAAPRVPSLRGAYR 106
+ + L D F +++LN +S + ++++ + ++ QLA +P L ++R
Sbjct 100 IPFVENLKPDERFKVKILLNENSRVGDTSLYSWTIDIISQLAVTTIPKLEFSFR 153
Lambda K H
0.321 0.139 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1608078824
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40