bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1089_orf1
Length=165
Score E
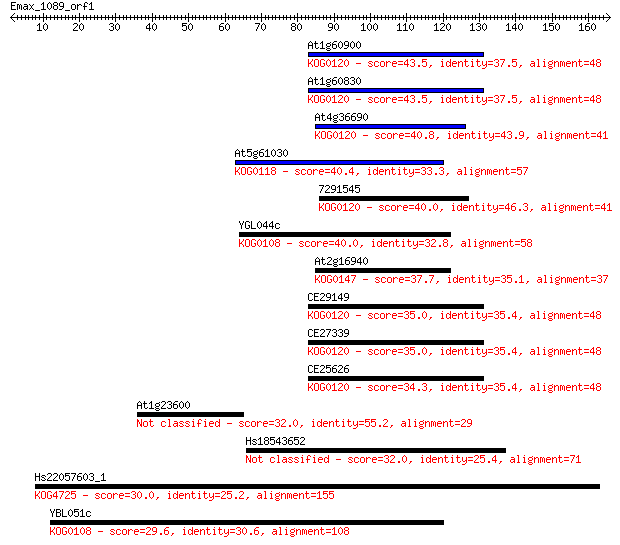
Sequences producing significant alignments: (Bits) Value
At1g60900 43.5 2e-04
At1g60830 43.5 2e-04
At4g36690 40.8 0.001
At5g61030 40.4 0.001
7291545 40.0 0.002
YGL044c 40.0 0.002
At2g16940 37.7 0.009
CE29149 35.0 0.066
CE27339 35.0 0.066
CE25626 34.3 0.12
At1g23600 32.0 0.55
Hs18543652 32.0 0.56
Hs22057603_1 30.0 2.3
YBL051c 29.6 2.5
> At1g60900
Length=568
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 29/48 (60%), Gaps = 0/48 (0%)
Query 83 PMIGYAYVAFVDCEASANARKALNGRKFGGRVVEAHYYSELNFLRRNF 130
P +G ++ + D + S+ AR +NGRKFGG V A YY E + + ++
Sbjct 519 PGVGKVFLEYADVDGSSKARSGMNGRKFGGNQVVAVYYPEDKYAQGDY 566
> At1g60830
Length=111
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 29/48 (60%), Gaps = 0/48 (0%)
Query 83 PMIGYAYVAFVDCEASANARKALNGRKFGGRVVEAHYYSELNFLRRNF 130
P +G ++ + D + S+ AR +NGRKFGG V A YY E + + ++
Sbjct 62 PGVGKVFLEYADVDGSSKARSGMNGRKFGGNQVVAVYYPEDKYAQGDY 109
> At4g36690
Length=573
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/41 (43%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 85 IGYAYVAFVDCEASANARKALNGRKFGGRVVEAHYYSELNF 125
+G ++ + D + S AR +NGRKFGG V A YY E F
Sbjct 526 LGKVFLKYADTDGSTRARFGMNGRKFGGNEVVAVYYPEDKF 566
> At5g61030
Length=309
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 35/57 (61%), Gaps = 6/57 (10%)
Query 63 EIVTEEMVLNAIEGKTKVLPPMIGYAYVAFVDCEASANARKALNGRKFGGRVVEAHY 119
E+V ++L+ G+++ G+ +V F EA+++A +AL+GR GRVV+ +Y
Sbjct 65 EVVDTRVILDRETGRSR------GFGFVTFTSSEAASSAIQALDGRDLHGRVVKVNY 115
> 7291545
Length=449
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/41 (46%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 86 GYAYVAFVDCEASANARKALNGRKFGGRVVEAHYYSELNFL 126
G +V F E S A KAL+GRKF GR+V YY +L
Sbjct 404 GKVFVQFESVEDSQKALKALSGRKFSGRIVMTSYYDPEKYL 444
> YGL044c
Length=296
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 34/58 (58%), Gaps = 6/58 (10%)
Query 64 IVTEEMVLNAIEGKTKVLPPMIGYAYVAFVDCEASANARKALNGRKFGGRVVEAHYYS 121
++ +M+ + G++K GYA++ F D E+SA+A + LNG + G R ++ Y S
Sbjct 44 VINLKMMFDPQTGRSK------GYAFIEFRDLESSASAVRNLNGYQLGSRFLKCGYSS 95
> At2g16940
Length=600
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 13/37 (35%), Positives = 26/37 (70%), Gaps = 0/37 (0%)
Query 85 IGYAYVAFVDCEASANARKALNGRKFGGRVVEAHYYS 121
+G+ Y+ F + +A+ A++AL+GR F G+++ A Y +
Sbjct 550 VGFVYLRFENAQAAIGAQRALHGRWFAGKMITATYMT 586
> CE29149
Length=474
Score = 35.0 bits (79), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 21/48 (43%), Gaps = 0/48 (0%)
Query 83 PMIGYAYVAFVDCEASANARKALNGRKFGGRVVEAHYYSELNFLRRNF 130
P +G +V F A+ AL GRKF R V YY + R F
Sbjct 427 PGVGKVFVEFASTSDCQRAQAALTGRKFANRTVVTSYYDVDKYHNRQF 474
> CE27339
Length=496
Score = 35.0 bits (79), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 21/48 (43%), Gaps = 0/48 (0%)
Query 83 PMIGYAYVAFVDCEASANARKALNGRKFGGRVVEAHYYSELNFLRRNF 130
P +G +V F A+ AL GRKF R V YY + R F
Sbjct 449 PGVGKVFVEFASTSDCQRAQAALTGRKFANRTVVTSYYDVDKYHNRQF 496
> CE25626
Length=143
Score = 34.3 bits (77), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 21/48 (43%), Gaps = 0/48 (0%)
Query 83 PMIGYAYVAFVDCEASANARKALNGRKFGGRVVEAHYYSELNFLRRNF 130
P +G +V F A+ AL GRKF R V YY + R F
Sbjct 96 PGVGKVFVEFASTSDCQRAQAALTGRKFANRTVVTSYYDVDKYHNRQF 143
> At1g23600
Length=270
Score = 32.0 bits (71), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 16/29 (55%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 36 KGESPGSSSSNSNSNNNNNNNNNDDDEEI 64
KGES S + SNSN + + NN +DDEEI
Sbjct 20 KGESKRSENVKSNSNTDMDTNNTEDDEEI 48
> Hs18543652
Length=291
Score = 32.0 bits (71), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 18/71 (25%), Positives = 34/71 (47%), Gaps = 2/71 (2%)
Query 66 TEEMVLNAIEGKTKVLPPMIGYAYVAFVDCEASANARKALNGRKFGGRVVEAHYYSELNF 125
T E +L A+ K ++ G+ ++ F + NA K +NG+ F G+ ++ +L+F
Sbjct 19 TNEKMLKAVFAKQGLI--SEGFVFIIFEKSADAKNAAKDMNGKSFDGKAIKVEQSQKLSF 76
Query 126 LRRNFSSPSPN 136
P P+
Sbjct 77 QSGGRWRPPPS 87
> Hs22057603_1
Length=651
Score = 30.0 bits (66), Expect = 2.3, Method: Composition-based stats.
Identities = 39/161 (24%), Positives = 64/161 (39%), Gaps = 6/161 (3%)
Query 8 RQVKRETTEGDTTTADGSSSSNKNGAGVKGESPGSSSSNSNSNNNNNNNNNDDDE-EIVT 66
R K + +T T+ G S + G+ GESP SS++ + NN E E+
Sbjct 34 RNRKTNGSIHETATSGGCHSPGDSATGIHGESPTSSATLKDLEEKKANNEKQKAERELEV 93
Query 67 EEMVLNAIEGK--TKVLPPMIGYAYVAFVDCEASANARKALNGRKFGGRVVEAHYYSELN 124
+ LN +GK T + Y + + + +L + R + A ++
Sbjct 94 QIQRLNIQKGKLNTDLYHTKRSLRYFEEESKDLAVRLQHSLQRKGELERALSAVTATQKK 153
Query 125 FLRRNFSSPSP---NFKKGHSYLHSPPLAAIVNKLKEEIKQ 162
R FSS S +K S L A + +LKE +K+
Sbjct 154 KAERQFSSRSKARTEWKLEQSMREQALLKAQLTQLKESLKE 194
> YBL051c
Length=668
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 33/136 (24%), Positives = 54/136 (39%), Gaps = 32/136 (23%)
Query 12 RETTEGDTTTADGSSSSNKNGAGVKGESPGSSSSNSNSNNNNNNNNND------DDEEI- 64
+ET + T D N GV+ E+ +S+ N+ N ++ N NN DD+ I
Sbjct 29 QETNQQSIETRDAIDKEN----GVQTETGENSAKNAEQNVSSTNLNNAPTNGALDDDVIP 84
Query 65 -----------VTEEMVLNAIEGKTKVLPPMIGY----------AYVAFVDCEASANARK 103
+ +E +L+ IE LP Y A+ F E +
Sbjct 85 NAIVIKNIPFAIKKEQLLDIIEEMDLPLPYAFNYHFDNGIFRGLAFANFTTPEETTQVIT 144
Query 104 ALNGRKFGGRVVEAHY 119
+LNG++ GR ++ Y
Sbjct 145 SLNGKEISGRKLKVEY 160
Lambda K H
0.304 0.125 0.343
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2353551590
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40