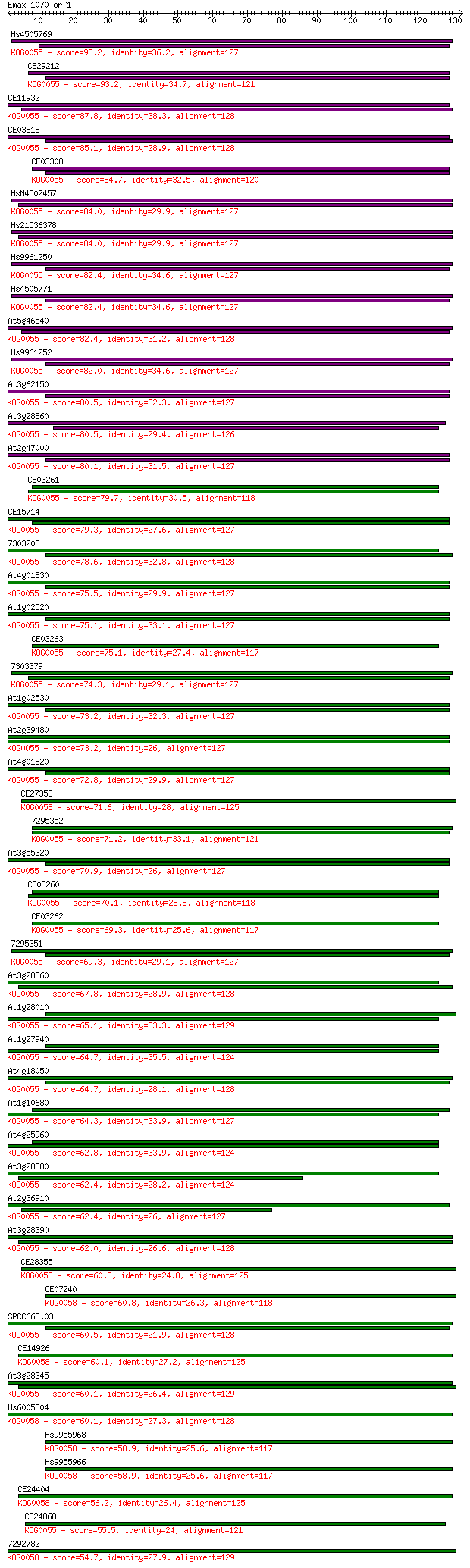
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1070_orf1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
Hs4505769 93.2 1e-19
CE29212 93.2 1e-19
CE11932 87.8 5e-18
CE03818 85.1 4e-17
CE03308 84.7 4e-17
HsM4502457 84.0 8e-17
Hs21536378 84.0 8e-17
Hs9961250 82.4 2e-16
Hs4505771 82.4 2e-16
At5g46540 82.4 2e-16
Hs9961252 82.0 2e-16
At3g62150 80.5 8e-16
At3g28860 80.5 8e-16
At2g47000 80.1 9e-16
CE03261 79.7 1e-15
CE15714 79.3 2e-15
7303208 78.6 3e-15
At4g01830 75.5 3e-14
At1g02520 75.1 3e-14
CE03263 75.1 3e-14
7303379 74.3 5e-14
At1g02530 73.2 1e-13
At2g39480 73.2 1e-13
At4g01820 72.8 1e-13
CE27353 71.6 4e-13
7295352 71.2 4e-13
At3g55320 70.9 5e-13
CE03260 70.1 1e-12
CE03262 69.3 2e-12
7295351 69.3 2e-12
At3g28360 67.8 5e-12
At1g28010 65.1 4e-11
At1g27940 64.7 4e-11
At4g18050 64.7 5e-11
At1g10680 64.3 5e-11
At4g25960 62.8 2e-10
At3g28380 62.4 2e-10
At2g36910 62.4 2e-10
At3g28390 62.0 3e-10
CE28355 60.8 5e-10
CE07240 60.8 7e-10
SPCC663.03 60.5 9e-10
CE14926 60.1 9e-10
At3g28345 60.1 1e-09
Hs6005804 60.1 1e-09
Hs9955968 58.9 2e-09
Hs9955966 58.9 2e-09
CE24404 56.2 2e-08
CE24868 55.5 2e-08
7292782 54.7 4e-08
> Hs4505769
Length=1279
Score = 93.2 bits (230), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 46/127 (36%), Positives = 72/127 (56%), Gaps = 0/127 (0%)
Query 2 QQAKAFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVL 61
+Q ++++F AI+RQE+ WFD +VG L +R+ +V+ I G+G K+ M Q
Sbjct 142 RQIHKIRKQFFHAIMRQEIGWFDVHDVGELNTRLTDDVSKINEGIGDKIGMFFQSMATFF 201
Query 62 GALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAI 121
IVGF + WKLTLV A PV+ + A + + +Y+ AG+V+EE + AI
Sbjct 202 TGFIVGFTRGWKLTLVILAISPVLGLSAAVWAKILSSFTDKELLAYAKAGAVAEEVLAAI 261
Query 122 RTVVSLA 128
RTV++
Sbjct 262 RTVIAFG 268
Score = 71.6 bits (174), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 43/123 (34%), Positives = 74/123 (60%), Gaps = 6/123 (4%)
Query 10 RY--FQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLG-AL 64
RY F++++RQ+++WFD ++ G+L +R+ + A ++ +G +LA++ Q + A LG +
Sbjct 788 RYMVFRSMLRQDVSWFDDPKNTTGALTTRLANDAAQVKGAIGSRLAVITQ-NIANLGTGI 846
Query 65 IVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTV 124
I+ FI W+LTL+ A VP++A G + + Q + AG ++ EAI RTV
Sbjct 847 IISFIYGWQLTLLLLAIVPIIAIAGVVEMKMLSGQALKDKKELEGAGKIATEAIENFRTV 906
Query 125 VSL 127
VSL
Sbjct 907 VSL 909
> CE29212
Length=1265
Score = 93.2 bits (230), Expect = 1e-19, Method: Composition-based stats.
Identities = 42/121 (34%), Positives = 74/121 (61%), Gaps = 0/121 (0%)
Query 7 FKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIV 66
++ Y +AI+RQ++ WFD+ G+L +R+ ++ +R G+G K A+L+Q+ A L V
Sbjct 146 LRQNYLKAILRQQIQWFDKQQTGNLTARLTDDLERVREGLGDKFALLVQMFAAFLAGYGV 205
Query 67 GFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTVVS 126
GF SW +TLV P++ G ++ ++ + + E+Y+ AG+++EE +IRTV S
Sbjct 206 GFFYSWSMTLVMMGFAPLIVLSGAKMSKSMATRTRVEQETYAVAGAIAEETFSSIRTVHS 265
Query 127 L 127
L
Sbjct 266 L 266
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 37/120 (30%), Positives = 65/120 (54%), Gaps = 7/120 (5%)
Query 12 FQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVGFI 69
F+ ++RQ++A++D + G L +R + ++R V +L +++ + GAL +GF
Sbjct 781 FKNLLRQDIAFYDDLRHGTGKLCTRFATDAPNVRY-VFTRLPVVLASIVTICGALGIGFY 839
Query 70 KSWKLTLVCAAAVPVVAGFGGMVAWAVH--KQEKDTMESYSSAGSVSEEAIMAIRTVVSL 127
W+L L+ VP++ GG + KQ +DT + AG V+ +A+ IRTV SL
Sbjct 840 YGWQLALILVVMVPLLV-MGGYFEMQMRFGKQIRDT-QLLEEAGKVASQAVEHIRTVHSL 897
> CE11932
Length=1321
Score = 87.8 bits (216), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 48/132 (36%), Positives = 77/132 (58%), Gaps = 10/132 (7%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAV 60
+Q + + ++I+RQE++WFD ++ G+LA+++ N+ ++ G G K+ M Q +
Sbjct 165 EQMNNRLRREFVKSILRQEISWFDTNHSGTLATKLFDNLERVKEGTGDKIGMAFQYLSQF 224
Query 61 LGALIVGFIKSWKLTLVCAAAVPVVA--GFG---GMVAWAVHKQEKDTMESYSSAGSVSE 115
+ IV F SW+LTLV A P+ A GF M +A+ ++T+ Y+ AG V E
Sbjct 225 ITGFIVAFTHSWQLTLVMLAVTPIQALCGFAIAKSMSTFAI----RETLR-YAKAGKVVE 279
Query 116 EAIMAIRTVVSL 127
E I +IRTVVSL
Sbjct 280 ETISSIRTVVSL 291
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 31/128 (24%), Positives = 70/128 (54%), Gaps = 6/128 (4%)
Query 5 KAFKERYFQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLG 62
+ + + F+ ++ Q + +FD Q+ G +++R+ +V ++R+ + + + +I +++
Sbjct 827 RDLRNKLFRNVLSQHIGFFDSPQNASGKISTRLATDVPNLRTAIDFRFSTVITTLVSMVA 886
Query 63 ALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWA--VHKQEKDTMESYSSAGSVSEEAIMA 120
+ + F W++ L+ A +P+VA FG + K K E ++ +G ++ EAI
Sbjct 887 GIGLAFFYGWQMALLIIAILPIVA-FGQYLRGRRFTGKNVKSASE-FADSGKIAIEAIEN 944
Query 121 IRTVVSLA 128
+RTV +LA
Sbjct 945 VRTVQALA 952
> CE03818
Length=1268
Score = 85.1 bits (209), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 37/127 (29%), Positives = 75/127 (59%), Gaps = 0/127 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAV 60
+++ +++Y ++++RQ+ WFD++ +G L +M + + I+ G+G K+ +L+
Sbjct 125 ERRLHCIRKKYLKSVLRQDAKWFDETTIGGLTQKMSSGIEKIKDGIGDKVGVLVGGVATF 184
Query 61 LGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMA 120
+ + +GF W+LTLV VP+ G + A +++ K+ M +YS+AG ++ E I
Sbjct 185 ISGVSIGFYMCWQLTLVMMITVPLQLGSMYLSAKHLNRATKNEMSAYSNAGGMANEVIAG 244
Query 121 IRTVVSL 127
IRTV++
Sbjct 245 IRTVMAF 251
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 29/119 (24%), Positives = 60/119 (50%), Gaps = 2/119 (1%)
Query 12 FQAIVRQEMAWFDQS--NVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVGFI 69
F+ I++Q+ ++FD S NVGSL SR+ + ++++ + +LA ++ ++ + V F
Sbjct 788 FRNIMQQDASYFDDSRHNVGSLTSRLATDAPNVQAAIDQRLAEVLTGIVSLFCGVGVAFY 847
Query 70 KSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTVVSLA 128
W + + A ++ VA + + + M+S A + E+I +TV +L
Sbjct 848 YGWNMAPIGLATALLLVVVQSSVAQYLKFRGQRDMDSAIEASRLVTESISNWKTVQALT 906
> CE03308
Length=1266
Score = 84.7 bits (208), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 39/120 (32%), Positives = 70/120 (58%), Gaps = 0/120 (0%)
Query 8 KERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVG 67
++ Q+++RQ+ WFD++ VG L +M + + I+ G+G K+ +L+ + + +G
Sbjct 130 RKHLLQSVLRQDAKWFDENTVGGLTQKMSSGIEKIKDGIGDKIGVLVSGIATFISGVALG 189
Query 68 FIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTVVSL 127
F W+LTLV VP+ G + A +++ K+ M +YSSAG ++ E I IRTV++
Sbjct 190 FYMCWQLTLVMLVTVPLQLGSMYLSAKHLNRATKNEMSAYSSAGGMANEVIAGIRTVIAF 249
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/119 (24%), Positives = 61/119 (51%), Gaps = 4/119 (3%)
Query 12 FQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVGFI 69
F+ I++Q+ +FD + NVG+L SR+ + ++++ + +LA ++ ++ + V F
Sbjct 786 FKNIMQQDATYFDDPKHNVGNLTSRLATDSQNVQAAIDHRLAEVLNGVVSLFTGIAVAFW 845
Query 70 KSWKLTLV-CAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTVVSL 127
W + + A+ +V + + ++ KD MES A + E+I +TV +L
Sbjct 846 FGWSMAPIGLITALLLVIAQSAVAQYLKYRGPKD-MESAIEASRIVTESISNWKTVQAL 903
> HsM4502457
Length=1321
Score = 84.0 bits (206), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 38/127 (29%), Positives = 73/127 (57%), Gaps = 0/127 (0%)
Query 2 QQAKAFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVL 61
+Q + ++ YF+ I+R E+ WFD ++VG L +R ++ I + ++A+ IQ T+ +
Sbjct 169 RQIQKMRKFYFRRIMRMEIGWFDCNSVGELNTRFSDDINKINDAIADQMALFIQRMTSTI 228
Query 62 GALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAI 121
++GF + WKLTLV + P++ + +V K +++Y+ AG V++E I ++
Sbjct 229 CGFLLGFFRGWKLTLVIISVSPLIGIGAATIGLSVSKFTDYELKAYAKAGVVADEVISSM 288
Query 122 RTVVSLA 128
RTV +
Sbjct 289 RTVAAFG 295
Score = 57.8 bits (138), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 34/127 (26%), Positives = 66/127 (51%), Gaps = 2/127 (1%)
Query 4 AKAFKERYFQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVL 61
K ++ F+A++ Q++AWFD +++ G+L +R+ + + ++ G ++ M++ T V
Sbjct 828 TKRLRKFGFRAMLGQDIAWFDDLRNSPGALTTRLATDASQVQGAAGSQIGMIVNSFTNVT 887
Query 62 GALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAI 121
A+I+ F SWKL+LV P +A G + ++ G ++ EA+ I
Sbjct 888 VAMIIAFSFSWKLSLVILCFFPFLALSGATQTRMLTGFASRDKQALEMVGQITNEALSNI 947
Query 122 RTVVSLA 128
RTV +
Sbjct 948 RTVAGIG 954
> Hs21536378
Length=1321
Score = 84.0 bits (206), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 38/127 (29%), Positives = 73/127 (57%), Gaps = 0/127 (0%)
Query 2 QQAKAFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVL 61
+Q + ++ YF+ I+R E+ WFD ++VG L +R ++ I + ++A+ IQ T+ +
Sbjct 169 RQIQKMRKFYFRRIMRMEIGWFDCNSVGELNTRFSDDINKINDAIADQMALFIQRMTSTI 228
Query 62 GALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAI 121
++GF + WKLTLV + P++ + +V K +++Y+ AG V++E I ++
Sbjct 229 CGFLLGFFRGWKLTLVIISVSPLIGIGAATIGLSVSKFTDYELKAYAKAGVVADEVISSM 288
Query 122 RTVVSLA 128
RTV +
Sbjct 289 RTVAAFG 295
Score = 57.8 bits (138), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 34/127 (26%), Positives = 66/127 (51%), Gaps = 2/127 (1%)
Query 4 AKAFKERYFQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVL 61
K ++ F+A++ Q++AWFD +++ G+L +R+ + + ++ G ++ M++ T V
Sbjct 828 TKRLRKFGFRAMLGQDIAWFDDLRNSPGALTTRLATDASQVQGAAGSQIGMIVNSFTNVT 887
Query 62 GALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAI 121
A+I+ F SWKL+LV P +A G + ++ G ++ EA+ I
Sbjct 888 VAMIIAFSFSWKLSLVILCFFPFLALSGATQTRMLTGFASRDKQALEMVGQITNEALSNI 947
Query 122 RTVVSLA 128
RTV +
Sbjct 948 RTVAGIG 954
> Hs9961250
Length=1286
Score = 82.4 bits (202), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 44/127 (34%), Positives = 74/127 (58%), Gaps = 0/127 (0%)
Query 2 QQAKAFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVL 61
+Q + ++++F AI+RQE+ WFD ++ L +R+ +++ I G+G K+ M Q
Sbjct 144 RQIRKIRQKFFHAILRQEIGWFDINDTTELNTRLTDDISKISEGIGDKVGMFFQAVATFF 203
Query 62 GALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAI 121
IVGFI+ WKLTLV A P++ + A + + +Y+ AG+V+EEA+ AI
Sbjct 204 AGFIVGFIRGWKLTLVIMAISPILGLSAAVWAKILSAFSDKELAAYAKAGAVAEEALGAI 263
Query 122 RTVVSLA 128
RTV++
Sbjct 264 RTVIAFG 270
Score = 77.8 bits (190), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 47/123 (38%), Positives = 80/123 (65%), Gaps = 12/123 (9%)
Query 12 FQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLG-ALIVGF 68
F+A++RQ+M+WFD +++ G+L++R+ + A ++ G +LA++ Q + A LG +I+ F
Sbjct 792 FKAMLRQDMSWFDDHKNSTGALSTRLATDAAQVQGATGTRLALIAQ-NIANLGTGIIISF 850
Query 69 IKSWKLTLVCAAAVPVVAGFGGMVAWAV----HKQEKDTMESYSSAGSVSEEAIMAIRTV 124
I W+LTL+ A VP++A G+V + K++K +E +AG ++ EAI IRTV
Sbjct 851 IYGWQLTLLLLAVVPIIA-VSGIVEMKLLAGNAKRDKKELE---AAGKIATEAIENIRTV 906
Query 125 VSL 127
VSL
Sbjct 907 VSL 909
> Hs4505771
Length=1279
Score = 82.4 bits (202), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 44/127 (34%), Positives = 74/127 (58%), Gaps = 0/127 (0%)
Query 2 QQAKAFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVL 61
+Q + ++++F AI+RQE+ WFD ++ L +R+ +++ I G+G K+ M Q
Sbjct 144 RQIRKIRQKFFHAILRQEIGWFDINDTTELNTRLTDDISKISEGIGDKVGMFFQAVATFF 203
Query 62 GALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAI 121
IVGFI+ WKLTLV A P++ + A + + +Y+ AG+V+EEA+ AI
Sbjct 204 AGFIVGFIRGWKLTLVIMAISPILGLSAAVWAKILSAFSDKELAAYAKAGAVAEEALGAI 263
Query 122 RTVVSLA 128
RTV++
Sbjct 264 RTVIAFG 270
Score = 77.8 bits (190), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 47/123 (38%), Positives = 80/123 (65%), Gaps = 12/123 (9%)
Query 12 FQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLG-ALIVGF 68
F+A++RQ+M+WFD +++ G+L++R+ + A ++ G +LA++ Q + A LG +I+ F
Sbjct 792 FKAMLRQDMSWFDDHKNSTGALSTRLATDAAQVQGATGTRLALIAQ-NIANLGTGIIISF 850
Query 69 IKSWKLTLVCAAAVPVVAGFGGMVAWAV----HKQEKDTMESYSSAGSVSEEAIMAIRTV 124
I W+LTL+ A VP++A G+V + K++K +E +AG ++ EAI IRTV
Sbjct 851 IYGWQLTLLLLAVVPIIA-VSGIVEMKLLAGNAKRDKKELE---AAGKIATEAIENIRTV 906
Query 125 VSL 127
VSL
Sbjct 907 VSL 909
> At5g46540
Length=1248
Score = 82.4 bits (202), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 40/129 (31%), Positives = 74/129 (57%), Gaps = 1/129 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFD-QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTA 59
++Q+ + Y + I+RQ++ +FD ++N G + RM + I+ +G K+ QL ++
Sbjct 105 ERQSTRIRRLYLKTILRQDIGFFDTETNTGEVIGRMSGDTILIQDSMGEKVGKFTQLVSS 164
Query 60 VLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIM 119
+G V FI KLTL VP++ G GG + + + K+ + +Y+ AG+V ++A+
Sbjct 165 FVGGFTVAFIVGMKLTLALLPCVPLIVGTGGAMTYIMSKKAQRVQLAYTEAGNVVQQAVG 224
Query 120 AIRTVVSLA 128
+IRTVV+
Sbjct 225 SIRTVVAFT 233
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 37/136 (27%), Positives = 67/136 (49%), Gaps = 24/136 (17%)
Query 5 KAFKERYFQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLG 62
K + F ++ Q+++WFD +++ G + +R+ + ++++S VG L +++Q ++G
Sbjct 756 KRIRSLSFDRVLHQDISWFDDTKNSSGVIGARLSTDASTVKSIVGDVLGLIMQNMATIIG 815
Query 63 ALIVGFIKSWKLTLVCAAAVPV-----------VAGFGGMVAWAVHKQEKDTMESYSSAG 111
A I+ F +W L L+ PV + GFG A A K Y A
Sbjct 816 AFIIAFTANWLLALMALLVAPVMFFQGYYQIKFITGFG---AKARGK--------YEEAS 864
Query 112 SVSEEAIMAIRTVVSL 127
V+ +A+ +IRTV S
Sbjct 865 QVASDAVSSIRTVASF 880
> Hs9961252
Length=1232
Score = 82.0 bits (201), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 44/127 (34%), Positives = 74/127 (58%), Gaps = 0/127 (0%)
Query 2 QQAKAFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVL 61
+Q + ++++F AI+RQE+ WFD ++ L +R+ +++ I G+G K+ M Q
Sbjct 144 RQIRKIRQKFFHAILRQEIGWFDINDTTELNTRLTDDISKISEGIGDKVGMFFQAVATFF 203
Query 62 GALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAI 121
IVGFI+ WKLTLV A P++ + A + + +Y+ AG+V+EEA+ AI
Sbjct 204 AGFIVGFIRGWKLTLVIMAISPILGLSAAVWAKILSAFSDKELAAYAKAGAVAEEALGAI 263
Query 122 RTVVSLA 128
RTV++
Sbjct 264 RTVIAFG 270
Score = 77.8 bits (190), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 47/123 (38%), Positives = 80/123 (65%), Gaps = 12/123 (9%)
Query 12 FQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLG-ALIVGF 68
F+A++RQ+M+WFD +++ G+L++R+ + A ++ G +LA++ Q + A LG +I+ F
Sbjct 792 FKAMLRQDMSWFDDHKNSTGALSTRLATDAAQVQGATGTRLALIAQ-NIANLGTGIIISF 850
Query 69 IKSWKLTLVCAAAVPVVAGFGGMVAWAV----HKQEKDTMESYSSAGSVSEEAIMAIRTV 124
I W+LTL+ A VP++A G+V + K++K +E +AG ++ EAI IRTV
Sbjct 851 IYGWQLTLLLLAVVPIIA-VSGIVEMKLLAGNAKRDKKELE---AAGKIATEAIENIRTV 906
Query 125 VSL 127
VSL
Sbjct 907 VSL 909
> At3g62150
Length=1292
Score = 80.5 bits (197), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 41/128 (32%), Positives = 71/128 (55%), Gaps = 1/128 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFD-QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTA 59
++QA + Y Q I+RQ++A+FD ++N G + RM + I+ +G K+ IQL +
Sbjct 151 ERQAGRIRSLYLQTILRQDIAFFDVETNTGEVVGRMSGDTVLIQDAMGEKVGKAIQLVST 210
Query 60 VLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIM 119
+G ++ F + W LTLV +++P++ G +A + K SY+ A V E+ +
Sbjct 211 FIGGFVIAFTEGWLLTLVMVSSIPLLVMSGAALAIVISKMASRGQTSYAKAAVVVEQTVG 270
Query 120 AIRTVVSL 127
+IRTV S
Sbjct 271 SIRTVASF 278
Score = 63.9 bits (154), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 38/124 (30%), Positives = 68/124 (54%), Gaps = 18/124 (14%)
Query 12 FQAIVRQEMAWFDQS--NVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVGFI 69
F+ +VR E+ WFD++ + G++ +R+ A+ A++R VG LA +Q +V L++ F+
Sbjct 810 FEKVVRMEVGWFDETENSSGAIGARLSADAATVRGLVGDALAQTVQNLASVTAGLVIAFV 869
Query 70 KSWKLTLVCAAAVPVVAGFGG------MVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRT 123
SW+L + A +P++ G G MV ++ +E A V+ +A+ +IRT
Sbjct 870 ASWQLAFIVLAMLPLI-GLNGYIYMKFMVGFSADAKE---------ASQVANDAVGSIRT 919
Query 124 VVSL 127
V S
Sbjct 920 VASF 923
> At3g28860
Length=1252
Score = 80.5 bits (197), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 37/127 (29%), Positives = 74/127 (58%), Gaps = 1/127 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFD-QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTA 59
++Q A +++Y +A+++Q++ +FD + G + + + ++ + K+ I +
Sbjct 113 ERQVAALRKKYLEAVLKQDVGFFDTDARTGDIVFSVSTDTLLVQDAISEKVGNFIHYLST 172
Query 60 VLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIM 119
L L+VGF+ +WKL L+ A +P +A GG+ A+ + + ESY++AG ++E+AI
Sbjct 173 FLAGLVVGFVSAWKLALLSVAVIPGIAFAGGLYAYTLTGITSKSRESYANAGVIAEQAIA 232
Query 120 AIRTVVS 126
+RTV S
Sbjct 233 QVRTVYS 239
Score = 57.8 bits (138), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 32/115 (27%), Positives = 66/115 (57%), Gaps = 6/115 (5%)
Query 14 AIVRQEMAWFDQS--NVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVGFIKS 71
AI+R E+ WFD+ N +A+R+ + A ++S + ++++++Q T++L + IV FI
Sbjct 770 AILRNEVGWFDEDEHNSSLIAARLATDAADVKSAIAERISVILQNMTSLLTSFIVAFIVE 829
Query 72 WKLTLVCAAAVP--VVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTV 124
W+++L+ P V+A F + ++ DT ++++ ++ E + IRTV
Sbjct 830 WRVSLLILGTFPLLVLANFAQQL--SLKGFAGDTAKAHAKTSMIAGEGVSNIRTV 882
> At2g47000
Length=1286
Score = 80.1 bits (196), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 40/128 (31%), Positives = 72/128 (56%), Gaps = 1/128 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFD-QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTA 59
++QA + Y + I+RQ++A+FD +N G + RM + I+ +G K+ IQL
Sbjct 132 ERQAARIRSLYLKTILRQDIAFFDIDTNTGEVVGRMSGDTVLIQDAMGEKVGKAIQLLAT 191
Query 60 VLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIM 119
+G ++ F++ W LTLV +++P++ G ++A + K +Y+ A +V E+ I
Sbjct 192 FVGGFVIAFVRGWLLTLVMLSSIPLLVMAGALLAIVIAKTASRGQTAYAKAATVVEQTIG 251
Query 120 AIRTVVSL 127
+IRTV S
Sbjct 252 SIRTVASF 259
Score = 70.1 bits (170), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 38/118 (32%), Positives = 67/118 (56%), Gaps = 2/118 (1%)
Query 12 FQAIVRQEMAWFDQ--SNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVGFI 69
F+ +V E+ WFD+ ++ G++ +R+ A+ A+IR VG LA +Q +++L LI+ F+
Sbjct 800 FEKVVHMEVGWFDEPENSSGTIGARLSADAATIRGLVGDSLAQTVQNLSSILAGLIIAFL 859
Query 70 KSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTVVSL 127
W+L V A +P++A G + + D + Y A V+ +A+ +IRTV S
Sbjct 860 ACWQLAFVVLAMLPLIALNGFLYMKFMKGFSADAKKMYGEASQVANDAVGSIRTVASF 917
> CE03261
Length=1291
Score = 79.7 bits (195), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 35/117 (29%), Positives = 69/117 (58%), Gaps = 0/117 (0%)
Query 8 KERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVG 67
+ RY +I+RQ WFD++ G++ +R+ N+ I+ GVG KL +LI+ + V+ ++++
Sbjct 172 RNRYISSILRQNAGWFDKNLSGTITTRLNDNMERIQDGVGDKLGVLIRGISMVIASVVIS 231
Query 68 FIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTV 124
I W+L L+ +PV +++ + K + +E AG+++EE +M +RT+
Sbjct 232 LIYEWRLALMMLGLIPVSTICMTLLSRFLEKSTGEELEKVGEAGAIAEECLMGVRTI 288
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/120 (22%), Positives = 64/120 (53%), Gaps = 3/120 (2%)
Query 7 FKERYFQAIVRQEMAWFDQSN--VGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGAL 64
F+ F+ ++ Q+ A+FD GSL +R+ A+ +++ V ++ ++ + AV+ +
Sbjct 806 FRVAAFRNLLYQDAAYFDNPAHAPGSLITRLAADPPCVKAVVDGRMMQVVYATAAVIACV 865
Query 65 IVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTV 124
+GFI W++ ++ A + ++ +A+ + + ME+ AG ++ E I ++T+
Sbjct 866 TIGFINCWQVAILGTALIFLLGFIMAGLAFKISIVAAEHMEN-DDAGKIAIEIIENVKTI 924
> CE15714
Length=1294
Score = 79.3 bits (194), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 35/127 (27%), Positives = 72/127 (56%), Gaps = 0/127 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAV 60
++ + F+ ++F +++RQE+AW+D++ G+L++++ N+ +R G G K+ + Q+
Sbjct 132 EKLSNRFRRQFFHSVMRQEIAWYDKNTSGTLSNKLFDNLERVREGTGDKVGLAFQMMAQF 191
Query 61 LGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMA 120
+G V F W LTL+ + P + G +A + + Y+ AG ++EE + +
Sbjct 192 IGGFAVAFTYDWLLTLIMMSLSPFMMICGLFLAKLLATAATKEAKQYAVAGGIAEEVLTS 251
Query 121 IRTVVSL 127
IRTV++
Sbjct 252 IRTVIAF 258
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 31/123 (25%), Positives = 61/123 (49%), Gaps = 4/123 (3%)
Query 8 KERYFQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALI 65
+ + ++ ++RQ+ +FD + + G + +R+ + +I+S + +L + +V G L
Sbjct 783 RSKVYRNVLRQDATYFDMPKHSPGRITTRLATDAPNIKSAIDYRLGSIFNAIASVGGGLG 842
Query 66 VGFIKSWKLTLVCAAAVPVVA-GFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTV 124
+ F W++ + A P +A G M+ + D E +AG + EAI IRTV
Sbjct 843 IAFYYGWQMAFLVMAIFPFMAVGQALMMKYHGGSATSDAKE-MENAGKTAMEAIENIRTV 901
Query 125 VSL 127
+L
Sbjct 902 QAL 904
> 7303208
Length=1313
Score = 78.6 bits (192), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 40/124 (32%), Positives = 73/124 (58%), Gaps = 0/124 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAV 60
Q + ++F++I+ Q+M W+D + G +ASRM +++ + G+ K+ M + A
Sbjct 173 HSQILTIRSKFFRSILHQDMKWYDFNQSGEVASRMNEDLSKMEDGLAEKVVMFVHYLVAF 232
Query 61 LGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMA 120
+G+L++ F+K W+L+LVC ++P+ G+VA A + K + Y+ A V+E A+
Sbjct 233 VGSLVLAFVKGWQLSLVCLTSLPLTFIAMGLVAVATSRLAKKEVTMYAGAAVVAEGALSG 292
Query 121 IRTV 124
IRTV
Sbjct 293 IRTV 296
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 35/120 (29%), Positives = 63/120 (52%), Gaps = 4/120 (3%)
Query 12 FQAIVRQEMAWFDQ--SNVGSLASRMEANVASIRSGVGLKLAMLIQ-LSTAVLGALIVGF 68
F+A++RQE+AWFD + GSL +R+ + A+++ G ++ ++Q +ST LG + +
Sbjct 828 FEAMLRQEVAWFDDKANGTGSLCARLSGDAAAVQGATGQRIGTIVQSISTLALG-IALSM 886
Query 69 IKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTVVSLA 128
W L LV A P + M + K+ + ++ + ++ E + IRTV SL
Sbjct 887 YYEWSLGLVALAFTPFILIAFYMQRTLMAKENMGSAKTMENCTKLAVEVVSNIRTVASLG 946
> At4g01830
Length=1230
Score = 75.5 bits (184), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 38/128 (29%), Positives = 69/128 (53%), Gaps = 1/128 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFD-QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTA 59
++QA + Y + I+RQ++ +FD + G + RM + I +G K+ IQL +
Sbjct 101 ERQAARIRSLYLKTILRQDIGFFDVEMTTGEVVGRMSGDTVLILDAMGEKVGKFIQLIST 160
Query 60 VLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIM 119
+G ++ F++ W LTLV ++P++A G +A V + +Y+ A +V E+ +
Sbjct 161 FVGGFVIAFLRGWLLTLVMLTSIPLLAMSGAAIAIIVTRASSQEQAAYAKASNVVEQTLG 220
Query 120 AIRTVVSL 127
+IRTV S
Sbjct 221 SIRTVASF 228
Score = 50.8 bits (120), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 33/119 (27%), Positives = 62/119 (52%), Gaps = 4/119 (3%)
Query 12 FQAIVRQEMAWFDQ--SNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVGFI 69
F+ +V E+ WFD+ ++ G++ +R+ A+ A IR+ VG L + ++ +++ LI+ F
Sbjct 743 FEKVVHMEVGWFDEPGNSSGAMGARLSADAALIRTLVGDSLCLSVKNVASLVTGLIIAFT 802
Query 70 KSWKLTLVCAAAVPVVAGFGGMVAWAVHKQ-EKDTMESYSSAGSVSEEAIMAIRTVVSL 127
SW++ ++ + G G + K D Y A V+ +A+ +IRTV S
Sbjct 803 ASWEVAIIILVII-PFIGINGYIQIKFMKGFSADAKAKYEEASQVANDAVGSIRTVASF 860
> At1g02520
Length=1278
Score = 75.1 bits (183), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 72/128 (56%), Gaps = 1/128 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFD-QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTA 59
++QA + Y + I+RQ++ +FD ++N G + RM + I+ +G K+ IQL +
Sbjct 129 ERQAARIRSTYLKTILRQDIGFFDVETNTGEVVGRMSGDTVLIQDAMGEKVGKFIQLVST 188
Query 60 VLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIM 119
+G ++ FIK W LTLV ++P++A G +A V + +Y+ A +V E+ I
Sbjct 189 FVGGFVLAFIKGWLLTLVMLTSIPLLAMAGAAMALIVTRASSRGQAAYAKAATVVEQTIG 248
Query 120 AIRTVVSL 127
+IRTV S
Sbjct 249 SIRTVASF 256
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 39/119 (32%), Positives = 63/119 (52%), Gaps = 4/119 (3%)
Query 12 FQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVGFI 69
F+ V E+AWFD Q++ G++ +R+ A+ IR+ VG L++ +Q + LI+ F
Sbjct 790 FEKAVHMEVAWFDEPQNSSGTMGARLSADATLIRALVGDALSLAVQNVASAASGLIIAFT 849
Query 70 KSWKLTLVCAAAVPVVAGFGGMVAWAVHKQ-EKDTMESYSSAGSVSEEAIMAIRTVVSL 127
SW+L L+ +P++ G G V K D Y A V+ +A+ +IRTV S
Sbjct 850 ASWELALIILVMLPLI-GINGFVQVKFMKGFSADAKSKYEEASQVANDAVGSIRTVASF 907
> CE03263
Length=1270
Score = 75.1 bits (183), Expect = 3e-14, Method: Composition-based stats.
Identities = 32/117 (27%), Positives = 66/117 (56%), Gaps = 0/117 (0%)
Query 8 KERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVG 67
+ RY +++RQ WFD+ + G++A+++ ++ IR G+G KL +L++ ++ +++V
Sbjct 178 RHRYVYSVLRQNAGWFDKHHSGTIATKLNDSMERIREGIGDKLGVLLRGVAMLVASVVVA 237
Query 68 FIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTV 124
+I W+L + P G ++A + + AGS++EE++M +RTV
Sbjct 238 YIYEWRLACMLLGVAPTCIGCMSLMARQMTATTVKELGGVEKAGSIAEESLMGVRTV 294
> 7303379
Length=1279
Score = 74.3 bits (181), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 37/127 (29%), Positives = 71/127 (55%), Gaps = 0/127 (0%)
Query 2 QQAKAFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVL 61
Q ++ + +A++RQ++AW+D S+ + AS+M ++ ++ G+G K+ +++ L +
Sbjct 146 NQIDRIRKLFLEAMLRQDIAWYDTSSGSNFASKMTEDLDKLKEGIGEKIVIVVFLIMTFV 205
Query 62 GALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAI 121
++ F+ WKLTLV + VP + +VA + ++SYS A +V EE I
Sbjct 206 IGIVSAFVYGWKLTLVVLSCVPFIIAATSVVARLQGSLAEKELKSYSDAANVVEEVFSGI 265
Query 122 RTVVSLA 128
RTV + +
Sbjct 266 RTVFAFS 272
Score = 35.8 bits (81), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 35/123 (28%), Positives = 69/123 (56%), Gaps = 2/123 (1%)
Query 7 FKERYFQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGAL 64
++R F I+ Q++A+FD +++VG+L SR+ ++ ++++ G ++ ++Q ++ +
Sbjct 788 LRKRAFGTIIGQDIAYFDDERNSVGALCSRLASDCSNVQGATGARVGTMLQAVATLVVGM 847
Query 65 IVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTV 124
+VGF+ SW+ TL+ +P+V + + K + S A V+ EAI IRTV
Sbjct 848 VVGFVFSWQQTLLTLVTLPLVCLSVYLEGRFIMKSAQKAKASIEEASQVAVEAITNIRTV 907
Query 125 VSL 127
L
Sbjct 908 NGL 910
> At1g02530
Length=1273
Score = 73.2 bits (178), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 41/128 (32%), Positives = 69/128 (53%), Gaps = 1/128 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFD-QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTA 59
++QA + Y + I+RQ++ +FD ++N G + RM + I+ +G K+ IQL +
Sbjct 116 ERQAAKIRSNYLKTILRQDIGFFDVETNTGEVVGRMSGDTVHIQDAMGEKVGKFIQLVST 175
Query 60 VLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIM 119
+G + F K W LTLV ++P +A G +A V + +Y+ A +V E+ I
Sbjct 176 FVGGFALAFAKGWLLTLVMLTSIPFLAMAGAAMALLVTRASSRGQAAYAKAATVVEQTIG 235
Query 120 AIRTVVSL 127
+IRTV S
Sbjct 236 SIRTVASF 243
Score = 61.2 bits (147), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 64/119 (53%), Gaps = 4/119 (3%)
Query 12 FQAIVRQEMAWFDQ--SNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVGFI 69
F+ V E++WFD+ ++ G++ +R+ + A IR+ VG L++ +Q + + LI+ F
Sbjct 785 FEKAVHMEVSWFDEPENSSGTMGARLSTDAALIRALVGDALSLAVQNAASAASGLIIAFT 844
Query 70 KSWKLTLVCAAAVPVVAGFGGMVAWAVHKQ-EKDTMESYSSAGSVSEEAIMAIRTVVSL 127
SW+L L+ +P++ G G + K D Y A V+ +A+ +IRTV S
Sbjct 845 ASWELALIILVMLPLI-GINGFLQVKFMKGFSADAKSKYEEASQVANDAVGSIRTVASF 902
> At2g39480
Length=1407
Score = 73.2 bits (178), Expect = 1e-13, Method: Composition-based stats.
Identities = 33/128 (25%), Positives = 72/128 (56%), Gaps = 1/128 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFDQ-SNVGSLASRMEANVASIRSGVGLKLAMLIQLSTA 59
++Q + +Y Q ++ Q+M++FD N G + S++ ++V I+S + K+ I
Sbjct 162 ERQTAVIRSKYVQVLLNQDMSFFDTYGNNGDIVSQVLSDVLLIQSALSEKVGNYIHNMAT 221
Query 60 VLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIM 119
+ LI+GF+ W++ L+ A P + GG+ +H+ ++ ++Y+ A S++E+A+
Sbjct 222 FISGLIIGFVNCWEIALITLATGPFIVAAGGISNIFLHRLAENIQDAYAEAASIAEQAVS 281
Query 120 AIRTVVSL 127
+RT+ +
Sbjct 282 YVRTLYAF 289
Score = 57.8 bits (138), Expect = 5e-09, Method: Composition-based stats.
Identities = 34/129 (26%), Positives = 67/129 (51%), Gaps = 2/129 (1%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFDQ--SNVGSLASRMEANVASIRSGVGLKLAMLIQLST 58
++ + + F A++R E+ W+D+ ++ +L+ R+ + +R+ +L++ IQ S
Sbjct 905 EKMTERVRRMMFSAMLRNEVGWYDEEENSPDTLSMRLANDATFVRAAFSNRLSIFIQDSF 964
Query 59 AVLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAI 118
AV+ A+++G + W+L LV A +PV+ + K E + A V E+A+
Sbjct 965 AVIVAILIGLLLGWRLALVALATLPVLTLSAIAQKLWLAGFSKGIQEMHRKASLVLEDAV 1024
Query 119 MAIRTVVSL 127
I TVV+
Sbjct 1025 RNIYTVVAF 1033
> At4g01820
Length=1229
Score = 72.8 bits (177), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 38/128 (29%), Positives = 69/128 (53%), Gaps = 1/128 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFD-QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTA 59
++QA + Y + I+RQ++ +FD +++ G + RM + I +G K+ IQL
Sbjct 96 ERQAARIRSLYLKTILRQDIGFFDVETSTGEVVGRMSGDTVLILEAMGEKVGKFIQLIAT 155
Query 60 VLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIM 119
+G ++ F+K W LTLV ++P++A G + V + +Y+ A +V E+ +
Sbjct 156 FVGGFVLAFVKGWLLTLVMLVSIPLLAIAGAAMPIIVTRASSREQAAYAKASTVVEQTLG 215
Query 120 AIRTVVSL 127
+IRTV S
Sbjct 216 SIRTVASF 223
Score = 65.5 bits (158), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 35/118 (29%), Positives = 65/118 (55%), Gaps = 2/118 (1%)
Query 12 FQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVGFI 69
F+ +V E+ WFD +++ G++ SR+ A+ A I++ VG L++ ++ + A + LI+ F
Sbjct 742 FEKVVHMEVGWFDDPENSSGTIGSRLSADAALIKTLVGDSLSLSVKNAAAAVSGLIIAFT 801
Query 70 KSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTVVSL 127
SWKL ++ +P++ G + + D Y A V+ +A+ +IRTV S
Sbjct 802 ASWKLAVIILVMIPLIGINGYLQIKFIKGFTADAKAKYEEASQVANDAVGSIRTVASF 859
> CE27353
Length=815
Score = 71.6 bits (174), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 35/125 (28%), Positives = 69/125 (55%), Gaps = 0/125 (0%)
Query 5 KAFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGAL 64
+ + F+++V+QE+ +FD + G + SR+ A+ ++ + + L + +L + T + G+L
Sbjct 277 RQIRNDLFRSVVKQEIGFFDMNKTGEICSRLSADCQTMSNTLSLYMNVLTRNLTMLFGSL 336
Query 65 IVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTV 124
I F SWKL+++ +P++ + ++T S + A V+EE + +IRTV
Sbjct 337 IFMFTLSWKLSMITLINIPIIFLVNKIFGVWYDMLSEETQNSVAKANDVAEEVLSSIRTV 396
Query 125 VSLAC 129
S AC
Sbjct 397 KSFAC 401
> 7295352
Length=1320
Score = 71.2 bits (173), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 40/121 (33%), Positives = 61/121 (50%), Gaps = 0/121 (0%)
Query 8 KERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVG 67
+ +F+A +RQE+ W D + + A R+ N+ IRSG+ L +++ V+ ++++
Sbjct 159 RREFFKATLRQEIGWHDMAKDQNFAVRITDNMEKIRSGIAENLGHYVEIMCDVIISVVLS 218
Query 68 FIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTVVSL 127
FI WKL L +P+ VA K SY A SV EE I AIRTVV+
Sbjct 219 FIYGWKLALAIVFYIPLTLVVNSAVAHYQGKLTGQEQSSYVRASSVVEEVIGAIRTVVAF 278
Query 128 A 128
Sbjct 279 G 279
Score = 67.8 bits (164), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 40/129 (31%), Positives = 71/129 (55%), Gaps = 16/129 (12%)
Query 8 KERYFQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALI 65
+ + F+ I+ QEM WFD ++++G+L++R+ + AS++ +G L+ +IQ T + ++
Sbjct 821 RSKTFRCIMNQEMGWFDRKENSIGALSARLSGDAASVQGAIGFPLSNIIQAFTNFICSIA 880
Query 66 VGFIKSWKLTLVCAAAVPVV-------AGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAI 118
+ F SW+L L+C + P + A FG A +EK+ +E S ++ E I
Sbjct 881 IAFPYSWELALICLSTSPFMVASIVFEARFGEKSAL----KEKEVLEETS---RIATETI 933
Query 119 MAIRTVVSL 127
IRTV L
Sbjct 934 TQIRTVAGL 942
> At3g55320
Length=1408
Score = 70.9 bits (172), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 33/128 (25%), Positives = 71/128 (55%), Gaps = 1/128 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFDQ-SNVGSLASRMEANVASIRSGVGLKLAMLIQLSTA 59
++Q + +Y Q ++ Q+M++FD N G + S++ ++V I+S + K+ I
Sbjct 164 ERQTAVIRSKYVQVLLNQDMSFFDTYGNNGDIVSQVLSDVLLIQSALSEKVGNYIHNMAT 223
Query 60 VLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIM 119
+ L++GF+ W++ L+ A P + GG+ +H+ ++ ++Y+ A ++E+AI
Sbjct 224 FISGLVIGFVNCWEIALITLATGPFIVAAGGISNIFLHRLAENIQDAYAEAAGIAEQAIS 283
Query 120 AIRTVVSL 127
IRT+ +
Sbjct 284 YIRTLYAF 291
Score = 58.2 bits (139), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 38/129 (29%), Positives = 65/129 (50%), Gaps = 24/129 (18%)
Query 12 FQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVGFI 69
F A++R E+ WFD +++ +L+ R+ + +R+ +L++ IQ S AV+ AL++G +
Sbjct 917 FSAMLRNEVGWFDDEENSPDTLSMRLANDATFVRAAFSNRLSIFIQDSFAVIVALLIGLL 976
Query 70 KSWKLTLVCAAAVPV-----------VAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAI 118
W+L LV A +P+ +AGF K E + A V E+A+
Sbjct 977 LGWRLALVALATLPILTLSAIAQKLWLAGFS-----------KGIQEMHRKASLVLEDAV 1025
Query 119 MAIRTVVSL 127
I TVV+
Sbjct 1026 RNIYTVVAF 1034
> CE03260
Length=1318
Score = 70.1 bits (170), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 33/117 (28%), Positives = 65/117 (55%), Gaps = 0/117 (0%)
Query 8 KERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVG 67
K+ Y Q+I+RQ WFD+++ G+L +++ N+ I G+G KL +LI+ + +++
Sbjct 172 KQHYIQSILRQNAGWFDKNHSGTLTTKLHDNMERINEGIGDKLGVLIRGMVMFVAGIVIS 231
Query 68 FIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTV 124
F W+L L+ P+ +++ ++ + AGS++EE++M +RTV
Sbjct 232 FFYEWRLALMMMGIGPLCCVCMSLMSRSMSSFTSKELAGVGKAGSIAEESLMGVRTV 288
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 25/122 (20%), Positives = 64/122 (52%), Gaps = 7/122 (5%)
Query 7 FKERYFQAIVRQEMAWFDQSN--VGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGAL 64
F+ + F+ ++ Q+ ++FD G L +R+ ++ ++++ V ++ +I T++ L
Sbjct 833 FRVQSFKNLLYQDASFFDNPAHAPGKLITRLASDAPNVKAVVDTRMLQVIYSITSITINL 892
Query 65 IVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESY--SSAGSVSEEAIMAIR 122
I G+I W++ + A ++ F M+ +K ++ ++ AG ++ E I +++
Sbjct 893 ITGYIFCWRIAI---AGTIMIVLFATMMISMAYKIARENLKQIRKDEAGKIAIEIIESVK 949
Query 123 TV 124
T+
Sbjct 950 TI 951
> CE03262
Length=1327
Score = 69.3 bits (168), Expect = 2e-12, Method: Composition-based stats.
Identities = 30/117 (25%), Positives = 65/117 (55%), Gaps = 0/117 (0%)
Query 8 KERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVG 67
+ R+ +++RQ WFD+++ G++ +++ ++ IR G+G KL +L++ ++ A++V
Sbjct 184 RHRFVYSVLRQNAGWFDKNHSGTITTKLNDSMERIREGIGDKLGVLLRGFAMLIAAIVVA 243
Query 68 FIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTV 124
+I W+L + P ++A + + AGS++EE++M +RTV
Sbjct 244 YIYEWRLASMMLGVAPTCCICMSLLARQMTSTTIKELIGVGKAGSIAEESLMGVRTV 300
> 7295351
Length=1302
Score = 69.3 bits (168), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/127 (29%), Positives = 65/127 (51%), Gaps = 0/127 (0%)
Query 2 QQAKAFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVL 61
+Q + + F +++RQ++ W D ++ + M +V IR G+ K+ + L +
Sbjct 148 RQVTRMRIKLFSSVIRQDIGWHDLASKQNFTQSMVDDVEKIRDGISEKVGHFVYLVVGFI 207
Query 62 GALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAI 121
+ + F WKLTL ++ +P+V VA K ESY+ AG+++EE + +I
Sbjct 208 ITVAISFSYGWKLTLAVSSYIPLVILLNYYVAKFQGKLTAREQESYAGAGNLAEEILSSI 267
Query 122 RTVVSLA 128
RTVVS
Sbjct 268 RTVVSFG 274
Score = 64.3 bits (155), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 41/122 (33%), Positives = 66/122 (54%), Gaps = 10/122 (8%)
Query 12 FQAIVRQEMAWFDQSN--VGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVGFI 69
F A+V QE+ WFD N VG+L++R+ I+ +G L+ +IQ + + ++ V
Sbjct 813 FNAMVNQEVGWFDDENNSVGALSARLSGEAVDIQGAIGYPLSGMIQALSNFISSVSVAMY 872
Query 70 KSWKLTLVCAAAVPVVAGF----GGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTVV 125
+WKL L+C A P++ G M++ AV + EK +E A ++ E+I IRTV
Sbjct 873 YNWKLALLCLANCPIIVGSVILEAKMMSNAVVR-EKQVIE---EACRIATESITNIRTVA 928
Query 126 SL 127
L
Sbjct 929 GL 930
> At3g28360
Length=1158
Score = 67.8 bits (164), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 75/126 (59%), Gaps = 2/126 (1%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFDQ--SNVGSLASRMEANVASIRSGVGLKLAMLIQLST 58
++QA +ERY +A++RQ++ +FD ++ + + + ++ I+ + KL ++ ++
Sbjct 24 ERQAAKMRERYLRAVLRQDVGYFDLHVTSTSDIITSVSSDSLVIQDFLSEKLPNILMNAS 83
Query 59 AVLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAI 118
A +G+ IVGF+ W+LT+V + ++ G M A+ + E Y+ AGS++E+AI
Sbjct 84 AFVGSYIVGFMLLWRLTIVGFPFIILLLIPGLMYGRALIGISRKIREEYNEAGSIAEQAI 143
Query 119 MAIRTV 124
++RTV
Sbjct 144 SSVRTV 149
Score = 52.8 bits (125), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 31/132 (23%), Positives = 70/132 (53%), Gaps = 12/132 (9%)
Query 4 AKAFKERYFQAIVRQEMAWFDQ--SNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVL 61
K +E+ I+ E+ WFD+ ++ G++ SR+ + +RS VG ++++L+Q + V+
Sbjct 661 TKRIREQMLSKILTFEVNWFDEEENSSGAICSRLAKDANVVRSLVGERMSLLVQTISTVM 720
Query 62 GALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHK-----QEKDTMESYSSAGSVSEE 116
A +G + +W+ T+V + PV+ +V + + + K + + + ++ E
Sbjct 721 VACTIGLVIAWRFTIVMISVQPVI-----IVCYYIQRVLLKNMSKKAIIAQDESSKLAAE 775
Query 117 AIMAIRTVVSLA 128
A+ IRT+ + +
Sbjct 776 AVSNIRTITTFS 787
> At1g28010
Length=1247
Score = 65.1 bits (157), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 41/131 (31%), Positives = 65/131 (49%), Gaps = 24/131 (18%)
Query 12 FQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVGFI 69
F AI+ E+ WFD ++N GSL S + A+ +RS + +L+ ++Q + + AL + F
Sbjct 764 FSAILSNEIGWFDLDENNTGSLTSILAADATLVRSAIADRLSTIVQNLSLTITALALAFF 823
Query 70 KSWKLTLVCAAAVPV-----------VAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAI 118
SW++ V A P+ + GFGG D +YS A S++ EAI
Sbjct 824 YSWRVAAVVTACFPLLIAASLTEQLFLKGFGG-----------DYTRAYSRATSLAREAI 872
Query 119 MAIRTVVSLAC 129
IRTV + +
Sbjct 873 SNIRTVAAFSA 883
Score = 55.5 bits (132), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 28/125 (22%), Positives = 66/125 (52%), Gaps = 1/125 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFD-QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTA 59
++Q + Y ++I+ +++ +FD ++ + + ++ ++ +G K +++
Sbjct 120 ERQTARLRINYLKSILAKDITFFDTEARDSNFIFHISSDAILVQDAIGDKTGHVLRYLCQ 179
Query 60 VLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIM 119
+ ++GF+ W+LTL+ VP++A GG A + + + +Y+ AG V+EE +
Sbjct 180 FIAGFVIGFLSVWQLTLLTLGVVPLIAIAGGGYAIVMSTISEKSEAAYADAGKVAEEVMS 239
Query 120 AIRTV 124
+RTV
Sbjct 240 QVRTV 244
> At1g27940
Length=1245
Score = 64.7 bits (156), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 42/126 (33%), Positives = 63/126 (50%), Gaps = 24/126 (19%)
Query 12 FQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVGFI 69
F AI+ E+ WFD ++N GSL S + A+ +RS + +L+ ++Q + + AL + F
Sbjct 762 FSAILSNEIGWFDLDENNTGSLTSILAADATLVRSALADRLSTIVQNLSLTVTALALAFF 821
Query 70 KSWKLTLVCAAAVPV-----------VAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAI 118
SW++ V A P+ + GFGG D +YS A SV+ EAI
Sbjct 822 YSWRVAAVVTACFPLLIAASLTEQLFLKGFGG-----------DYTRAYSRATSVAREAI 870
Query 119 MAIRTV 124
IRTV
Sbjct 871 ANIRTV 876
Score = 58.2 bits (139), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 29/125 (23%), Positives = 68/125 (54%), Gaps = 1/125 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFD-QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTA 59
++Q + Y ++I+ +++ +FD ++ +L + ++ ++ +G K +++ +
Sbjct 119 ERQTARLRINYLKSILAKDITFFDTEARDSNLIFHISSDAILVQDAIGDKTDHVLRYLSQ 178
Query 60 VLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIM 119
+ ++GF+ W+LTL+ VP++A GG A + + + +Y+ AG V+EE +
Sbjct 179 FIAGFVIGFLSVWQLTLLTLGVVPLIAIAGGGYAIVMSTISEKSETAYADAGKVAEEVMS 238
Query 120 AIRTV 124
+RTV
Sbjct 239 QVRTV 243
> At4g18050
Length=1323
Score = 64.7 bits (156), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 36/129 (27%), Positives = 66/129 (51%), Gaps = 1/129 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFD-QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTA 59
++Q+ + Y + I+RQ++ +FD ++N G + RM + I+ +G K+ QL
Sbjct 103 ERQSATIRGLYLKTILRQDIGYFDTETNTGEVIGRMSGDTILIQDAMGEKVGKFTQLLCT 162
Query 60 VLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIM 119
LG + F K L V + +P++ G ++ + K +Y+ AG+V E+ +
Sbjct 163 FLGGFAIAFYKGPLLAGVLCSCIPLIVIAGAAMSLIMSKMAGRGQVAYAEAGNVVEQTVG 222
Query 120 AIRTVVSLA 128
AIRTVV+
Sbjct 223 AIRTVVAFT 231
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 36/145 (24%), Positives = 58/145 (40%), Gaps = 29/145 (20%)
Query 12 FQAIVRQEMAWFDQSNVGSLAS-------------------------RME----ANVASI 42
F +V QE++WFD + + R+E + +++
Sbjct 766 FDKVVHQEISWFDDTANSRYYNFIYIINRRILYVLILIFICVLLPPVRLERECSTDASTV 825
Query 43 RSGVGLKLAMLIQLSTAVLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKD 102
RS VG LA+++Q V LI+ F +W L L+ A P + G + D
Sbjct 826 RSLVGDALALIVQNIATVTTGLIIAFTANWILALIVLALSPFIVIQGYAQTKFLTGFSAD 885
Query 103 TMESYSSAGSVSEEAIMAIRTVVSL 127
Y A V+ +A+ +IRTV S
Sbjct 886 AKAMYEEASQVANDAVSSIRTVASF 910
> At1g10680
Length=1227
Score = 64.3 bits (155), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 41/124 (33%), Positives = 72/124 (58%), Gaps = 6/124 (4%)
Query 8 KERYFQAIVRQEMAWFDQ-SNVGS-LASRMEANVASIRSGVGLKLAMLIQLSTAVLGALI 65
+++ F AI+R E+ WFD+ N S LASR+E++ +R+ V + +L++ V+ A I
Sbjct 738 RQKMFSAILRNEIGWFDKVDNTSSMLASRLESDATLLRTIVVDRSTILLENLGLVVTAFI 797
Query 66 VGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEK--DTMESYSSAGSVSEEAIMAIRT 123
+ FI +W+LTLV A P++ G ++ + Q + ++Y A ++ E+I IRT
Sbjct 798 ISFILNWRLTLVVLATYPLI--ISGHISEKIFMQGYGGNLSKAYLKANMLAGESISNIRT 855
Query 124 VVSL 127
VV+
Sbjct 856 VVAF 859
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 29/125 (23%), Positives = 65/125 (52%), Gaps = 1/125 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFD-QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTA 59
++QA ++ Y ++++ Q+++ FD + + G + S + + + ++ + K+ + +
Sbjct 113 ERQAAKIRKAYLRSMLSQDISLFDTEISTGEVISAITSEILVVQDAISEKVGNFMHFISR 172
Query 60 VLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIM 119
+ +GF W+++LV + VP +A GG+ A+ +SY A ++EE I
Sbjct 173 FIAGFAIGFASVWQISLVTLSIVPFIALAGGIYAFVSSGLIVRVRKSYVKANEIAEEVIG 232
Query 120 AIRTV 124
+RTV
Sbjct 233 NVRTV 237
> At4g25960
Length=1233
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 40/121 (33%), Positives = 70/121 (57%), Gaps = 6/121 (4%)
Query 8 KERYFQAIVRQEMAWFDQ-SNVGS-LASRMEANVASIRSGVGLKLAMLIQLSTAVLGALI 65
+E F+AI++ E+ WFD+ N S LASR+E++ +++ V + +L+Q V+ + I
Sbjct 746 RENMFRAILKNEIGWFDEVDNTSSMLASRLESDATLLKTIVVDRSTILLQNLGLVVTSFI 805
Query 66 VGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEK--DTMESYSSAGSVSEEAIMAIRT 123
+ FI +W+LTLV A P+V G ++ + Q D ++Y A ++ E++ IRT
Sbjct 806 IAFILNWRLTLVVLATYPLV--ISGHISEKLFMQGYGGDLNKAYLKANMLAGESVSNIRT 863
Query 124 V 124
V
Sbjct 864 V 864
Score = 57.8 bits (138), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 30/125 (24%), Positives = 68/125 (54%), Gaps = 1/125 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFD-QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTA 59
++QA + Y ++++ Q+++ FD +++ G + S + +++ ++ + K+ + +
Sbjct 109 ERQAAKMRRAYLRSMLSQDISLFDTEASTGEVISAITSDILVVQDALSEKVGNFLHYISR 168
Query 60 VLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIM 119
+ +GF W+++LV + VP++A GG+ A+ +SY AG ++EE I
Sbjct 169 FIAGFAIGFTSVWQISLVTLSIVPLIALAGGIYAFVAIGLIARVRKSYIKAGEIAEEVIG 228
Query 120 AIRTV 124
+RTV
Sbjct 229 NVRTV 233
> At3g28380
Length=1240
Score = 62.4 bits (150), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 35/126 (27%), Positives = 72/126 (57%), Gaps = 2/126 (1%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFDQ--SNVGSLASRMEANVASIRSGVGLKLAMLIQLST 58
++QA +E+Y +A++RQ++ +FD ++ + + + ++ I+ + KL + ++
Sbjct 107 ERQAARMREKYLRAVLRQDVGYFDLHVTSTSDVITSISSDSLVIQDFLSEKLPNFLMNAS 166
Query 59 AVLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAI 118
A + + IV FI W+LT+V + ++ G M A+ + E Y+ AGS++E+AI
Sbjct 167 AFVASYIVSFILMWRLTIVGFPFIILLLVPGLMYGRALVSISRKIHEQYNEAGSIAEQAI 226
Query 119 MAIRTV 124
++RTV
Sbjct 227 SSVRTV 232
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 25/84 (29%), Positives = 51/84 (60%), Gaps = 2/84 (2%)
Query 4 AKAFKERYFQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVL 61
K +E+ I+ E+ WFD ++ G++ SR+ + +RS VG ++++L+Q +AV+
Sbjct 745 TKRIREQMLSKILTFEVNWFDIDDNSSGAICSRLAKDANVVRSMVGDRMSLLVQTISAVI 804
Query 62 GALIVGFIKSWKLTLVCAAAVPVV 85
A I+G + +W+L +V + P++
Sbjct 805 IACIIGLVIAWRLAIVMISVQPLI 828
> At2g36910
Length=1286
Score = 62.4 bits (150), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 33/128 (25%), Positives = 63/128 (49%), Gaps = 1/128 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFD-QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTA 59
++Q + +Y +A + Q++ +FD + + + + ++ + KL I
Sbjct 116 ERQTTKMRIKYLEAALNQDIQFFDTEVRTSDVVFAINTDAVMVQDAISEKLGNFIHYMAT 175
Query 60 VLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIM 119
+ IVGF W+L LV A VP++A GG+ + K + ES S AG++ E+ ++
Sbjct 176 FVSGFIVGFTAVWQLALVTLAVVPLIAVIGGIHTTTLSKLSNKSQESLSQAGNIVEQTVV 235
Query 120 AIRTVVSL 127
IR V++
Sbjct 236 QIRVVMAF 243
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 46/74 (62%), Gaps = 2/74 (2%)
Query 5 KAFKERYFQAIVRQEMAWFDQSNVGS--LASRMEANVASIRSGVGLKLAMLIQLSTAVLG 62
K +E+ A+++ EMAWFDQ S +A+R+ + ++RS +G ++++++Q + +L
Sbjct 774 KRVREKMLSAVLKNEMAWFDQEENESARIAARLALDANNVRSAIGDRISVIVQNTALMLV 833
Query 63 ALIVGFIKSWKLTL 76
A GF+ W+L L
Sbjct 834 ACTAGFVLQWRLAL 847
> At3g28390
Length=1225
Score = 62.0 bits (149), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 34/130 (26%), Positives = 73/130 (56%), Gaps = 2/130 (1%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFDQ--SNVGSLASRMEANVASIRSGVGLKLAMLIQLST 58
++QA +E+Y +A++RQ++ +FD ++ + + + ++ I+ + KL + ++
Sbjct 95 ERQAAKMREKYLKAVLRQDVGYFDLHVTSTSDVITSVSSDSLVIQDFLSEKLPNFLMNTS 154
Query 59 AVLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAI 118
A + + IVGF+ W+LT+V + ++ G M A+ + E Y+ AGS++E+ I
Sbjct 155 AFVASYIVGFLLLWRLTIVGFPFIILLLIPGLMYGRALIRISMKIREEYNEAGSIAEQVI 214
Query 119 MAIRTVVSLA 128
++RTV +
Sbjct 215 SSVRTVYAFG 224
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 30/127 (23%), Positives = 66/127 (51%), Gaps = 2/127 (1%)
Query 4 AKAFKERYFQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVL 61
K +ER I+ E+ WFD +++ G++ SR+ + +RS VG ++++L+Q +AV
Sbjct 730 TKRIRERMLGKILTFEVNWFDKDENSSGAICSRLAKDANMVRSLVGDRMSLLVQTISAVS 789
Query 62 GALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAI 121
+G + SW+ ++V + PV+ + ++ ++ + ++ EA+ I
Sbjct 790 ITCAIGLVISWRFSIVMMSVQPVIVVCFYTQRVLLKSMSRNAIKGQDESSKLAAEAVSNI 849
Query 122 RTVVSLA 128
RT+ + +
Sbjct 850 RTITAFS 856
> CE28355
Length=787
Score = 60.8 bits (146), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 31/125 (24%), Positives = 65/125 (52%), Gaps = 0/125 (0%)
Query 5 KAFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGAL 64
+A + F +V+Q++A++D G + SR+ A+ ++ V L + + ++ +LG++
Sbjct 253 RAIRYDLFHGLVKQDVAFYDAHKTGEVTSRLAADCQTMSDTVALNVNVFLRNCVMLLGSM 312
Query 65 IVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTV 124
I SW+L+LV VP++ + + T ++ + + V+EE + +RTV
Sbjct 313 IFMMKLSWRLSLVTFILVPIIFVASKIFGTYYDLLSERTQDTIAESNDVAEEVLSTMRTV 372
Query 125 VSLAC 129
S +C
Sbjct 373 RSFSC 377
> CE07240
Length=761
Score = 60.8 bits (146), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 31/118 (26%), Positives = 62/118 (52%), Gaps = 0/118 (0%)
Query 12 FQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVGFIKS 71
F +++RQ++A++D + G SR+ ++ +I S V + + ++ ++GAL + S
Sbjct 277 FTSLIRQDIAFYDTAKTGDTMSRLTSDCQTISSTVSTNVNVFMRNGVMLIGALAFMILMS 336
Query 72 WKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTVVSLAC 129
W+L +V AVP+V + K + ++ + ++EE + +RTV S AC
Sbjct 337 WRLAMVTFIAVPLVGFITKAYSSFYDKISEKLQQTIAETNQMAEEVVSTMRTVRSFAC 394
> SPCC663.03
Length=1362
Score = 60.5 bits (145), Expect = 9e-10, Method: Composition-based stats.
Identities = 28/128 (21%), Positives = 61/128 (47%), Gaps = 0/128 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAV 60
++ A+ ++ Y AI+ Q + +FD+ G + +R+ + I+ G+G K+ ++
Sbjct 169 ERIARRIRQDYLHAILSQNIGYFDRLGAGEITTRITTDTNFIQDGLGEKVGLVFFAIATF 228
Query 61 LGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMA 120
+ ++ FI+ WK TL+ ++ P + G G+ + K K + + + + EE
Sbjct 229 VSGFVIAFIRHWKFTLILSSMFPAICGGIGLGVPFITKNTKGQIAVVAESSTFVEEVFSN 288
Query 121 IRTVVSLA 128
IR +
Sbjct 289 IRNAFAFG 296
Score = 56.6 bits (135), Expect = 1e-08, Method: Composition-based stats.
Identities = 32/118 (27%), Positives = 60/118 (50%), Gaps = 2/118 (1%)
Query 12 FQAIVRQEMAWFDQS--NVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVGFI 69
F+ ++RQ++ +FD+S VG++ + + + S+ G L Q+ T ++ I+
Sbjct 876 FRTLLRQDVEFFDRSENTVGAITTSLSTKIQSLEGLSGPTLGTFFQILTNIISVTILSLA 935
Query 70 KSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTVVSL 127
WKL LV + PV+ G A+ + ++ +Y + + + E+ AIRTV SL
Sbjct 936 TGWKLGLVTLSTSPVIITAGYYRVRALDQVQEKLSAAYKESAAFACESTSAIRTVASL 993
> CE14926
Length=633
Score = 60.1 bits (144), Expect = 9e-10, Method: Composition-based stats.
Identities = 34/125 (27%), Positives = 64/125 (51%), Gaps = 0/125 (0%)
Query 4 AKAFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGA 63
A+ + + F++I+ QE+++FD++ G L SR+ + SI + + + + L+ G+
Sbjct 127 ARNIRIQLFRSILHQEISFFDENKTGQLVSRITHDSESISNTLPVHIETLVNNLFMFFGS 186
Query 64 LIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRT 123
+ + F SW+LTL P+ AV K + ++ +++ EE + AIRT
Sbjct 187 IPIMFCYSWQLTLTSFIEFPITLLMTRYYGLAVEKLSEKENDATAASNETVEEVLSAIRT 246
Query 124 VVSLA 128
V S A
Sbjct 247 VRSFA 251
> At3g28345
Length=1240
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 34/130 (26%), Positives = 72/130 (55%), Gaps = 2/130 (1%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFDQ--SNVGSLASRMEANVASIRSGVGLKLAMLIQLST 58
++Q +E+Y +A++RQ++ +FD ++ + + + ++ I+ + KL + ++
Sbjct 107 ERQTARMREKYLRAVLRQDVGYFDLHVTSTSDVITSVSSDSFVIQDVLSEKLPNFLMSAS 166
Query 59 AVLGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAI 118
+G+ IVGFI W+L +V + ++ G M A+ + E Y+ AG V+E+AI
Sbjct 167 TFVGSYIVGFILLWRLAIVGLPFIVLLVIPGLMYGRALISISRKIREEYNEAGFVAEQAI 226
Query 119 MAIRTVVSLA 128
++RTV + +
Sbjct 227 SSVRTVYAFS 236
Score = 54.7 bits (130), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 33/133 (24%), Positives = 70/133 (52%), Gaps = 12/133 (9%)
Query 4 AKAFKERYFQAIVRQEMAWFD--QSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVL 61
K +ER ++ E+ WFD +++ G++ SR+ + +RS VG ++A+++Q +AV
Sbjct 745 TKRIRERMLSKVLTFEVGWFDRDENSSGAICSRLAKDANVVRSLVGDRMALVVQTVSAVT 804
Query 62 GALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHK-----QEKDTMESYSSAGSVSEE 116
A +G + +W+L LV A PV+ +V + + K +++ + ++ E
Sbjct 805 IAFTMGLVIAWRLALVMIAVQPVI-----IVCFYTRRVLLKSMSKKAIKAQDESSKLAAE 859
Query 117 AIMAIRTVVSLAC 129
A+ +RT+ + +
Sbjct 860 AVSNVRTITAFSS 872
> Hs6005804
Length=718
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 35/128 (27%), Positives = 67/128 (52%), Gaps = 0/128 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAV 60
++ A + F +++RQ + +FD + G L SR+ +V +S L ++ ++ + V
Sbjct 204 ERMAVDMRRALFSSLLRQNITFFDANKTGQLVSRLTTDVQEFKSSFKLVISQGLRSCSQV 263
Query 61 LGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMA 120
G L+ + S +LTL+ A P + G G ++ + K + E + A V++EA+
Sbjct 264 AGCLVSLSMLSTRLTLLLMVATPALMGVGTLMGSGLRKLSRQCQEHIARAMGVADEALGN 323
Query 121 IRTVVSLA 128
+RTV +LA
Sbjct 324 VRTVRALA 331
> Hs9955968
Length=723
Score = 58.9 bits (141), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/117 (25%), Positives = 61/117 (52%), Gaps = 0/117 (0%)
Query 12 FQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVGFIKS 71
F+++V QE ++FD++ G L SR+ ++ + V + + ++ + V G ++ F S
Sbjct 266 FRSLVSQETSFFDENRTGDLISRLTSDTTMVSDLVSQNINVFLRNTVKVTGVVVFMFSLS 325
Query 72 WKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTVVSLA 128
W+L+LV P++ + + K+ + + A + +EE I A++TV S A
Sbjct 326 WQLSLVTFMGFPIIMMVSNIYGKYYKRLSKEVQNALARASNTAEETISAMKTVRSFA 382
> Hs9955966
Length=766
Score = 58.9 bits (141), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/117 (25%), Positives = 61/117 (52%), Gaps = 0/117 (0%)
Query 12 FQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALIVGFIKS 71
F+++V QE ++FD++ G L SR+ ++ + V + + ++ + V G ++ F S
Sbjct 266 FRSLVSQETSFFDENRTGDLISRLTSDTTMVSDLVSQNINVFLRNTVKVTGVVVFMFSLS 325
Query 72 WKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTVVSLA 128
W+L+LV P++ + + K+ + + A + +EE I A++TV S A
Sbjct 326 WQLSLVTFMGFPIIMMVSNIYGKYYKRLSKEVQNALARASNTAEETISAMKTVRSFA 382
> CE24404
Length=807
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 33/128 (25%), Positives = 65/128 (50%), Gaps = 6/128 (4%)
Query 4 AKAFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGA 63
A+ + F+AI+ Q++++FD + G L SR+ + SI + + + + + S + G+
Sbjct 303 ARNIRVSLFRAILHQDISFFDDNKTGQLMSRITKDSESISNSLPVYIETTVNNSFMLFGS 362
Query 64 LIVGFIKSWKLTLVCAAAVPVV---AGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMA 120
+ F SW+L + P++ F G++ + ++E D + + + EE + A
Sbjct 363 APIMFYYSWQLAISTFVTFPIILLTTKFYGLIVEKLSEKEND---ATAVSNETVEEVLSA 419
Query 121 IRTVVSLA 128
IRTV S A
Sbjct 420 IRTVRSFA 427
> CE24868
Length=1239
Score = 55.5 bits (132), Expect = 2e-08, Method: Composition-based stats.
Identities = 29/121 (23%), Positives = 61/121 (50%), Gaps = 0/121 (0%)
Query 6 AFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAVLGALI 65
+ ++ + A++RQ+ W D+ + GS+ ++ N+ I G+G K ML++ ++I
Sbjct 154 SIRKEFLGAVLRQDANWLDKHSSGSITCQLNENIEVISDGLGNKCCMLVRGFAMFTSSII 213
Query 66 VGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMAIRTVV 125
+W+LT + PV A ++ ++ M + + ++ EE+I+ +RTV
Sbjct 214 ACAFINWQLTFITFTMGPVSAFVLHLLTKVNEVSNEELMSLSAQSHAIIEESILNVRTVQ 273
Query 126 S 126
S
Sbjct 274 S 274
> 7292782
Length=761
Score = 54.7 bits (130), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 36/129 (27%), Positives = 64/129 (49%), Gaps = 0/129 (0%)
Query 1 QQQAKAFKERYFQAIVRQEMAWFDQSNVGSLASRMEANVASIRSGVGLKLAMLIQLSTAV 60
+Q A ++ F IV Q++A+FD++ G L +R+ A+V ++ ++ ++ + +
Sbjct 211 EQMAAKMRQDLFTQIVVQDIAFFDENRTGELVNRLTADVQDFKTSFKQFVSQGLRSAAQL 270
Query 61 LGALIVGFIKSWKLTLVCAAAVPVVAGFGGMVAWAVHKQEKDTMESYSSAGSVSEEAIMA 120
+G I F+ S + + A+VP V F + + K + A V EEA+
Sbjct 271 IGGSISLFMISPHMAAIALASVPCVVMFMTYLGKKLRSLSKSSQAQAERATGVCEEALSN 330
Query 121 IRTVVSLAC 129
IRTV S AC
Sbjct 331 IRTVRSSAC 339
Lambda K H
0.322 0.129 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1283105810
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40