bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1068_orf1
Length=169
Score E
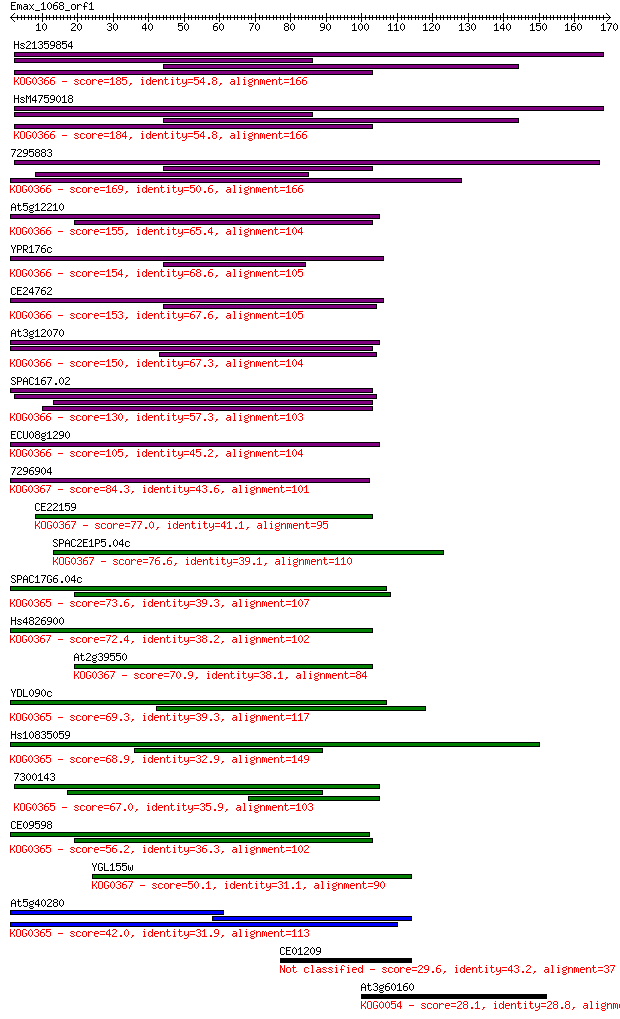
Sequences producing significant alignments: (Bits) Value
Hs21359854 185 3e-47
HsM4759018 184 6e-47
7295883 169 2e-42
At5g12210 155 4e-38
YPR176c 154 1e-37
CE24762 153 1e-37
At3g12070 150 8e-37
SPAC167.02 130 1e-30
ECU08g1290 105 3e-23
7296904 84.3 1e-16
CE22159 77.0 2e-14
SPAC2E1P5.04c 76.6 2e-14
SPAC17G6.04c 73.6 2e-13
Hs4826900 72.4 4e-13
At2g39550 70.9 1e-12
YDL090c 69.3 3e-12
Hs10835059 68.9 5e-12
7300143 67.0 2e-11
CE09598 56.2 3e-08
YGL155w 50.1 2e-06
At5g40280 42.0 6e-04
CE01209 29.6 3.0
At3g60160 28.1 9.4
> Hs21359854
Length=331
Score = 185 bits (470), Expect = 3e-47, Method: Compositional matrix adjust.
Identities = 91/166 (54%), Positives = 109/166 (65%), Gaps = 35/166 (21%)
Query 2 LSIAGQLHRIDVDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWINK 61
L+I QLH+++ D LGWWL ERQ+ SGGLNGRPEKLPD+CYSWWVL+SL IIGRLHWI++
Sbjct 201 LAITSQLHQVNSDLLGWWLCERQLPSGGLNGRPEKLPDVCYSWWVLASLKIIGRLHWIDR 260
Query 62 RKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLSHDYGQDLEVPAAADAAST 121
KL FILACQD ETGGFADRPG+MVDPFHT+FGIAGLSLL
Sbjct 261 EKLRNFILACQDEETGGFADRPGDMVDPFHTLFGIAGLSLLGE----------------- 303
Query 122 SMDVADETSDKIDVNKISEFIRKVNPVFCMLDSVIERRAIHIQRLS 167
E I+ VNPVFCM + V++R + + +S
Sbjct 304 ------------------EQIKPVNPVFCMPEEVLQRVNVQPELVS 331
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/84 (29%), Positives = 49/84 (58%), Gaps = 1/84 (1%)
Query 2 LSIAGQLHRIDVDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWINK 61
L++ ++ IDV+++ ++ Q G G D +S+ +++LA++G+L IN
Sbjct 105 LTLYDSINVIDVNKVVEYVKGLQKEDGSFAGDIWGEIDTRFSFCAVATLALLGKLDAINV 164
Query 62 RKLLQFILACQDPETGGFADRPGN 85
K ++F+L+C + + GGF RPG+
Sbjct 165 EKAIEFVLSCMNFD-GGFGCRPGS 187
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 53/100 (53%), Gaps = 26/100 (26%)
Query 44 WWVLSSLAIIGRLHWINKRKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLS 103
+W L+ + ++G+LH +N+ ++L FI +CQ E GG + G+ DP H ++ ++ + +L+
Sbjct 51 YWGLTVMDLMGQLHRMNREEILAFIKSCQH-ECGGISASIGH--DP-HLLYTLSAVQILT 106
Query 104 HDYGQDLEVPAAADAASTSMDVADETSDKIDVNKISEFIR 143
S++V IDVNK+ E+++
Sbjct 107 ---------------LYDSINV-------IDVNKVVEYVK 124
Score = 37.4 bits (85), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 24/101 (23%), Positives = 50/101 (49%), Gaps = 1/101 (0%)
Query 2 LSIAGQLHRIDVDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWINK 61
+ + GQLHR++ +++ ++ Q GG++ P L Y+ + L + ++ I+
Sbjct 57 MDLMGQLHRMNREEILAFIKSCQHECGGISASIGHDPHLLYTLSAVQILTLYDSINVIDV 116
Query 62 RKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLL 102
K+++++ Q E G FA +D + +A L+LL
Sbjct 117 NKVVEYVKGLQK-EDGSFAGDIWGEIDTRFSFCAVATLALL 156
> HsM4759018
Length=331
Score = 184 bits (468), Expect = 6e-47, Method: Compositional matrix adjust.
Identities = 91/166 (54%), Positives = 108/166 (65%), Gaps = 35/166 (21%)
Query 2 LSIAGQLHRIDVDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWINK 61
L+I QLH++ D LGWWL ERQ+ SGGLNGRPEKLPD+CYSWWVL+SL IIGRLHWI++
Sbjct 201 LAITSQLHQVTSDLLGWWLCERQLPSGGLNGRPEKLPDVCYSWWVLASLKIIGRLHWIDR 260
Query 62 RKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLSHDYGQDLEVPAAADAAST 121
KL FILACQD ETGGFADRPG+MVDPFHT+FGIAGLSLL
Sbjct 261 EKLRNFILACQDEETGGFADRPGDMVDPFHTLFGIAGLSLLGE----------------- 303
Query 122 SMDVADETSDKIDVNKISEFIRKVNPVFCMLDSVIERRAIHIQRLS 167
E I+ VNPVFCM + V++R + + +S
Sbjct 304 ------------------EQIKPVNPVFCMPEEVLQRVNVQPELVS 331
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 25/84 (29%), Positives = 49/84 (58%), Gaps = 1/84 (1%)
Query 2 LSIAGQLHRIDVDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWINK 61
L++ ++ IDV+++ ++ Q G G D +S+ +++LA++G+L IN
Sbjct 105 LTLYDSINVIDVNKVVEYVKGLQKEDGSFAGDIWGEIDTRFSFCAVATLALLGKLDAINV 164
Query 62 RKLLQFILACQDPETGGFADRPGN 85
K ++F+L+C + + GGF RPG+
Sbjct 165 EKAIEFVLSCMNSD-GGFGCRPGS 187
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 53/100 (53%), Gaps = 26/100 (26%)
Query 44 WWVLSSLAIIGRLHWINKRKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLS 103
+W L+ + ++G+LH +N+ ++L FI +CQ E GG + G+ DP H ++ ++ + +L+
Sbjct 51 YWGLTVMDLMGQLHRMNREEILAFIKSCQH-ECGGISASIGH--DP-HLLYTLSAVQILT 106
Query 104 HDYGQDLEVPAAADAASTSMDVADETSDKIDVNKISEFIR 143
S++V IDVNK+ E+++
Sbjct 107 ---------------LYDSINV-------IDVNKVVEYVK 124
Score = 37.4 bits (85), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 24/101 (23%), Positives = 50/101 (49%), Gaps = 1/101 (0%)
Query 2 LSIAGQLHRIDVDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWINK 61
+ + GQLHR++ +++ ++ Q GG++ P L Y+ + L + ++ I+
Sbjct 57 MDLMGQLHRMNREEILAFIKSCQHECGGISASIGHDPHLLYTLSAVQILTLYDSINVIDV 116
Query 62 RKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLL 102
K+++++ Q E G FA +D + +A L+LL
Sbjct 117 NKVVEYVKGLQK-EDGSFAGDIWGEIDTRFSFCAVATLALL 156
> 7295883
Length=347
Score = 169 bits (429), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 84/165 (50%), Positives = 101/165 (61%), Gaps = 35/165 (21%)
Query 2 LSIAGQLHRIDVDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWINK 61
S+ +LH +DVD+LGWWL ERQ+ SGGLNGRPEKLPD+CYSWWVL+SL I+GRLHWI+
Sbjct 215 FSLTHRLHLLDVDKLGWWLCERQLPSGGLNGRPEKLPDVCYSWWVLASLTIMGRLHWISS 274
Query 62 RKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLSHDYGQDLEVPAAADAAST 121
KL QFIL+CQD ETGGF+DR GNM D FHT+FGI GLSLL H
Sbjct 275 EKLQQFILSCQDTETGGFSDRTGNMPDIFHTLFGIGGLSLLGHSG--------------- 319
Query 122 SMDVADETSDKIDVNKISEFIRKVNPVFCMLDSVIERRAIHIQRL 166
++ +NP CM +I+R I Q L
Sbjct 320 --------------------LKAINPTLCMPQYIIDRLGIKPQLL 344
Score = 42.0 bits (97), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 38/59 (64%), Gaps = 3/59 (5%)
Query 44 WWVLSSLAIIGRLHWINKRKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLL 102
+W ++L I+G+L + ++ +++F+ CQ P TGGFA G+ DP H ++ ++ + +L
Sbjct 62 YWGTTALDIMGQLERLERKSIIEFVKRCQCPNTGGFAPCEGH--DP-HLLYTLSAIQIL 117
Score = 37.0 bits (84), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 40/78 (51%), Gaps = 1/78 (1%)
Query 8 LHRIDVDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRL-HWINKRKLLQ 66
L ID + + ++ Q G G D +S+ ++SL ++GR+ I+ K ++
Sbjct 123 LEEIDREAVVRFVVGLQQPDGSFFGDKWGEVDTRFSFCAVASLTLLGRMEQTIDVEKAVK 182
Query 67 FILACQDPETGGFADRPG 84
F+L+C + GGF +PG
Sbjct 183 FVLSCCNQTDGGFGSKPG 200
Score = 35.4 bits (80), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 31/128 (24%), Positives = 56/128 (43%), Gaps = 3/128 (2%)
Query 1 SLSIAGQLHRIDVDQLGWWLAERQMA-SGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWI 59
+L I GQL R++ + ++ Q +GG P L Y+ + L L I
Sbjct 67 ALDIMGQLERLERKSIIEFVKRCQCPNTGGFAPCEGHDPHLLYTLSAIQILCTYDALEEI 126
Query 60 NKRKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLSHDYGQDLEVPAAADAA 119
++ +++F++ Q P+ F D+ G VD + +A L+LL Q ++V A
Sbjct 127 DREAVVRFVVGLQQPDGSFFGDKWGE-VDTRFSFCAVASLTLLGR-MEQTIDVEKAVKFV 184
Query 120 STSMDVAD 127
+ + D
Sbjct 185 LSCCNQTD 192
> At5g12210
Length=313
Score = 155 bits (392), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 68/104 (65%), Positives = 84/104 (80%), Gaps = 0/104 (0%)
Query 1 SLSIAGQLHRIDVDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWIN 60
+L+I G LH +D D LGWWL ERQ+ +GGLNGRPEKL D+CYSWWVLSSL +I R+HWI+
Sbjct 186 ALAITGSLHHVDKDSLGWWLCERQLKAGGLNGRPEKLADVCYSWWVLSSLIMIDRVHWID 245
Query 61 KRKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLSH 104
K KL++FIL CQD + GG +DRP + VD FHT FG+AGLSLL +
Sbjct 246 KAKLVKFILDCQDLDNGGISDRPEDAVDIFHTYFGVAGLSLLEY 289
Score = 31.6 bits (70), Expect = 0.68, Method: Compositional matrix adjust.
Identities = 21/84 (25%), Positives = 38/84 (45%), Gaps = 1/84 (1%)
Query 19 WLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWINKRKLLQFILACQDPETGG 78
WL Q SGG G P + Y+ + LA+ +++ ++ K+ ++ Q+ E G
Sbjct 60 WLMTCQHESGGFAGNTGHDPHILYTLSAVQILALFDKINILDIGKVSSYVAKLQN-EDGS 118
Query 79 FADRPGNMVDPFHTVFGIAGLSLL 102
F+ +D + I LS+L
Sbjct 119 FSGDMWGEIDTRFSYIAICCLSIL 142
> YPR176c
Length=325
Score = 154 bits (388), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 72/108 (66%), Positives = 84/108 (77%), Gaps = 3/108 (2%)
Query 1 SLSIAGQLHRIDVDQL---GWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLH 57
+L+IA +L + DQL GWWL ERQ+ GGLNGRP KLPD+CYSWWVLSSLAIIGRL
Sbjct 193 ALAIANKLDMLSDDQLEEIGWWLCERQLPEGGLNGRPSKLPDVCYSWWVLSSLAIIGRLD 252
Query 58 WINKRKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLSHD 105
WIN KL +FIL CQD + GG +DRP N VD FHTVFG+AGLSL+ +D
Sbjct 253 WINYEKLTEFILKCQDEKKGGISDRPENEVDVFHTVFGVAGLSLMGYD 300
Score = 30.8 bits (68), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 12/40 (30%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 44 WWVLSSLAIIGRLHWINKRKLLQFILACQDPETGGFADRP 83
+W L++L ++ K +++ F+L+C D + G FA P
Sbjct 40 YWGLTALCVLDSPETFVKEEVISFVLSCWDDKYGAFAPFP 79
> CE24762
Length=335
Score = 153 bits (387), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 71/105 (67%), Positives = 84/105 (80%), Gaps = 0/105 (0%)
Query 1 SLSIAGQLHRIDVDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWIN 60
+L+IAG+L ID D+ WLA RQ SGGLNGRPEKLPD+CYSWWVL+SLAI+GRL++I+
Sbjct 206 ALAIAGRLDEIDRDRTAEWLAFRQCDSGGLNGRPEKLPDVCYSWWVLASLAILGRLNFID 265
Query 61 KRKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLSHD 105
+ +FI ACQD ETGGFADRPG+ DPFHTVFGIA LSL D
Sbjct 266 SDAMKKFIYACQDDETGGFADRPGDCADPFHTVFGIAALSLFGDD 310
Score = 31.2 bits (69), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 11/60 (18%), Positives = 34/60 (56%), Gaps = 1/60 (1%)
Query 44 WWVLSSLAIIGRLHWINKRKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLS 103
+W ++++ + +L ++ +++ ++L C++ + GG+ PG+ HT+ + L + +
Sbjct 57 YWCVNAMDLSKQLERMSTEEIVNYVLGCRNTD-GGYGPAPGHDSHLLHTLCAVQTLIIFN 115
> At3g12070
Length=317
Score = 150 bits (380), Expect = 8e-37, Method: Compositional matrix adjust.
Identities = 70/105 (66%), Positives = 82/105 (78%), Gaps = 1/105 (0%)
Query 1 SLSIAGQLHRIDVDQLGWWLAERQ-MASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWI 59
+L+I G LHR+D D LGWWL ERQ SGGLNGRPEKLPD+CYSWWVLSSL +I R+HWI
Sbjct 189 ALAITGNLHRVDKDLLGWWLCERQDYESGGLNGRPEKLPDVCYSWWVLSSLIMIDRVHWI 248
Query 60 NKRKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLSH 104
K KL++FIL QD + GG +DRP VD FHT FG+AGLSLL +
Sbjct 249 EKAKLVKFILDSQDMDNGGISDRPSYTVDIFHTYFGVAGLSLLEY 293
Score = 37.4 bits (85), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 26/102 (25%), Positives = 48/102 (47%), Gaps = 1/102 (0%)
Query 1 SLSIAGQLHRIDVDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWIN 60
+L++ +L + D++ W+ Q SGG G P + Y+ + LA+ +L+ ++
Sbjct 45 TLALLDKLGSVSEDEVVSWVMTCQHESGGFAGNTGHDPHVLYTLSAVQILALFDKLNILD 104
Query 61 KRKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLL 102
K+ +I Q+ E G F+ VD + I LS+L
Sbjct 105 VEKVSNYIAGLQN-EDGSFSGDIWGEVDTRFSYIAICCLSIL 145
Score = 36.6 bits (83), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 17/61 (27%), Positives = 42/61 (68%), Gaps = 4/61 (6%)
Query 43 SWWVLSSLAIIGRLHWINKRKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLL 102
++W L++LA++ +L +++ +++ +++ CQ E+GGFA G+ DP H ++ ++ + +L
Sbjct 39 AYWGLTTLALLDKLGSVSEDEVVSWVMTCQH-ESGGFAGNTGH--DP-HVLYTLSAVQIL 94
Query 103 S 103
+
Sbjct 95 A 95
> SPAC167.02
Length=311
Score = 130 bits (327), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 59/102 (57%), Positives = 76/102 (74%), Gaps = 0/102 (0%)
Query 1 SLSIAGQLHRIDVDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWIN 60
+L I +L ID + LGWW++ERQ+ GGLNGRPEKLPD CY WW LS LAIIG+L WI+
Sbjct 186 ALKILNKLDLIDEELLGWWISERQVKGGGLNGRPEKLPDSCYGWWDLSPLAIIGKLDWID 245
Query 61 KRKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLL 102
+ +L+ F+L QD ++GGFADR + D +HT F +AGLSLL
Sbjct 246 RNQLIDFLLGTQDADSGGFADRKEDATDVYHTCFSLAGLSLL 287
Score = 37.4 bits (85), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 25/102 (24%), Positives = 50/102 (49%), Gaps = 1/102 (0%)
Query 2 LSIAGQLHRIDVDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWINK 61
L++ LH +D D++ ++ Q G + G D + + ++ LAI+G+L ++NK
Sbjct 91 LAMLDSLHVVDKDKVASYIIGLQNEDGSMKGDRWGEIDARFLYSGINCLAILGKLDYLNK 150
Query 62 RKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLS 103
+ +++ C + + GGF PG +A L +L+
Sbjct 151 NTAVDWLMKCYNFD-GGFGLCPGAESHGAMVFTCVAALKILN 191
Score = 36.2 bits (82), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 24/90 (26%), Positives = 43/90 (47%), Gaps = 15/90 (16%)
Query 13 VDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWINKRKLLQFILACQ 72
D+L +WL E S +W S ++ + I+K +++ F+L+C
Sbjct 20 TDELDFWLKEHLHVSA--------------IYWSCMSFWLLKKKDQIDKERIVSFLLSCL 65
Query 73 DPETGGFADRPGNMVDPFHTVFGIAGLSLL 102
E+GGFA PG+ +TV+ + L++L
Sbjct 66 -TESGGFACYPGHDDHITNTVYAVQVLAML 94
Score = 29.3 bits (64), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 44/93 (47%), Gaps = 1/93 (1%)
Query 10 RIDVDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWINKRKLLQFIL 69
+ID +++ +L SGG P + + + + LA++ LH ++K K+ +I+
Sbjct 51 QIDKERIVSFLLSCLTESGGFACYPGHDDHITNTVYAVQVLAMLDSLHVVDKDKVASYII 110
Query 70 ACQDPETGGFADRPGNMVDPFHTVFGIAGLSLL 102
Q+ + DR G +D GI L++L
Sbjct 111 GLQNEDGSMKGDRWGE-IDARFLYSGINCLAIL 142
> ECU08g1290
Length=358
Score = 105 bits (263), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 47/104 (45%), Positives = 73/104 (70%), Gaps = 1/104 (0%)
Query 1 SLSIAGQLHRIDVDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWIN 60
+L G L +D +++ ++A +Q +SGGL+GR K D+CYS+W SSL +IG+ ++N
Sbjct 244 TLRSLGALETVDREEVARFIATKQASSGGLSGRVSKKEDVCYSFWAYSSLVLIGKECYVN 303
Query 61 KRKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLSH 104
+ +L +FI +CQ P +GGF+DRPGN D +H +F +AGLSLL +
Sbjct 304 QEELTRFIFSCQGP-SGGFSDRPGNETDLYHLMFALAGLSLLGY 346
> 7296904
Length=395
Score = 84.3 bits (207), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 44/104 (42%), Positives = 61/104 (58%), Gaps = 5/104 (4%)
Query 1 SLSIAGQLHRID---VDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLH 57
+L ++GQLHR+D V+++ WL RQM G GRP K D CYS+W+ +SL I+
Sbjct 220 ALHLSGQLHRLDATTVERMKRWLIFRQM--DGFQGRPNKPVDTCYSFWIGASLCILDGFE 277
Query 58 WINKRKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSL 101
+ + +FIL+ QD GGFA P DPFHT G+ GL+
Sbjct 278 LTDYARNREFILSTQDKLIGGFAKWPQATPDPFHTYLGLCGLAF 321
> CE22159
Length=360
Score = 77.0 bits (188), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 39/97 (40%), Positives = 56/97 (57%), Gaps = 4/97 (4%)
Query 8 LHRIDVDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWINKRKLLQF 67
L R D+D+L W ++Q G +GR K D CY++W+ ++L I+ H ++K+ L +F
Sbjct 238 LTRRDIDRLIRWAIQKQDI--GFHGRAHKPDDSCYAFWIGATLKILNAYHLVSKQHLREF 295
Query 68 ILACQDPETGGFAD--RPGNMVDPFHTVFGIAGLSLL 102
++ CQ P GGF PG D HT F IA LSLL
Sbjct 296 LMICQHPHIGGFCKYPEPGGYSDILHTYFSIAALSLL 332
> SPAC2E1P5.04c
Length=355
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 43/111 (38%), Positives = 62/111 (55%), Gaps = 6/111 (5%)
Query 13 VDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWINKRKLLQFILACQ 72
V++L WLA RQ++SGGLNGR K D CY++WVLSSL ++ L +I+ +L +++L
Sbjct 234 VERLIRWLASRQLSSGGLNGRTNKDVDTCYAYWVLSSLKLLDALPFIDGGELEKYLLLHA 293
Query 73 DPETGGFADRPGNMVDPFHTVFGIAGLSLLSHDYGQDLEVPAA-ADAASTS 122
GGF+ PG D H+ G+ ++ Y D P AD TS
Sbjct 294 QHALGGFSKTPGEFPDVLHSALGLYAMA-----YQDDKSFPKVNADIHMTS 339
> SPAC17G6.04c
Length=382
Score = 73.6 bits (179), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 41/118 (34%), Positives = 63/118 (53%), Gaps = 12/118 (10%)
Query 1 SLSIAGQLHRIDVDQLGWWLAERQ-MASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLH-- 57
++++ G L ++ +L WL +RQ A G +GR KL D CYSWWV +S I+ +
Sbjct 214 AIALLGGLDNLNEIKLSTWLVQRQDPALYGFSGRSNKLVDGCYSWWVGASHVIVASGYGS 273
Query 58 ---------WINKRKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLSHDY 106
+ N KLL +IL C +GG D+P D +HT + + GLS +++DY
Sbjct 274 ASHKSLPNLFYNPEKLLGYILQCCQSTSGGLRDKPPKRPDQYHTCYCLLGLSSIAYDY 331
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 27/89 (30%), Positives = 49/89 (55%), Gaps = 0/89 (0%)
Query 19 WLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWINKRKLLQFILACQDPETGG 78
WL + Q GGL+G P Y++ L+++A++G L +N+ KL +++ QDP G
Sbjct 184 WLCKCQTYEGGLSGVPYAEAHGGYTFCALAAIALLGGLDNLNEIKLSTWLVQRQDPALYG 243
Query 79 FADRPGNMVDPFHTVFGIAGLSLLSHDYG 107
F+ R +VD ++ + A +++ YG
Sbjct 244 FSGRSNKLVDGCYSWWVGASHVIVASGYG 272
> Hs4826900
Length=377
Score = 72.4 bits (176), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 39/106 (36%), Positives = 58/106 (54%), Gaps = 6/106 (5%)
Query 1 SLSIAGQLHRI----DVDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRL 56
SL + G+L + +++++ W RQ G +GRP K D CYS+WV ++L ++
Sbjct 229 SLCLMGKLEEVFSEKELNRIKRWCIMRQ--QNGYHGRPNKPVDTCYSFWVGATLKLLKIF 286
Query 57 HWINKRKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLL 102
+ N K +IL+ QD GGFA P + D H FGI GLSL+
Sbjct 287 QYTNFEKNRNYILSTQDRLVGGFAKWPDSHPDALHAYFGICGLSLM 332
> At2g39550
Length=375
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 32/84 (38%), Positives = 49/84 (58%), Gaps = 1/84 (1%)
Query 19 WLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWINKRKLLQFILACQDPETGG 78
W +RQ GG GR K D CY++W+ + L +IG I+K L +F+++CQ + GG
Sbjct 271 WCLQRQANDGGFQGRTNKPSDTCYAFWIGAVLKLIGGDALIDKMALRKFLMSCQS-KYGG 329
Query 79 FADRPGNMVDPFHTVFGIAGLSLL 102
F+ PG + D +H+ +G SLL
Sbjct 330 FSKFPGQLPDLYHSYYGYTAFSLL 353
> YDL090c
Length=431
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 42/111 (37%), Positives = 60/111 (54%), Gaps = 5/111 (4%)
Query 1 SLSIAGQLHRIDVDQLGWWLAERQMASG-GLNGRPEKLPDLCYSWWVLSSLAII---GRL 56
SL+I + +I+V++L W + RQ+ G GR KL D CYS+WV S AI+ G
Sbjct 268 SLAILRSMDQINVEKLLEWSSARQLQEERGFCGRSNKLVDGCYSFWVGGSAAILEAFGYG 327
Query 57 HWINKRKLLQFIL-ACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLSHDY 106
NK L +IL CQ+ E G D+PG D +HT + + GL++ Y
Sbjct 328 QCFNKHALRDYILYCCQEKEQPGLRDKPGAHSDFYHTNYCLLGLAVAESSY 378
Score = 34.3 bits (77), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 38/77 (49%), Gaps = 1/77 (1%)
Query 42 YSWWVLSSLAIIGRLHWINKRKLLQFILACQDPETGGFADRPGNMVDPFHTVF-GIAGLS 100
Y++ +SLAI+ + IN KLL++ A Q E GF R +VD ++ + G +
Sbjct 261 YTFCATASLAILRSMDQINVEKLLEWSSARQLQEERGFCGRSNKLVDGCYSFWVGGSAAI 320
Query 101 LLSHDYGQDLEVPAAAD 117
L + YGQ A D
Sbjct 321 LEAFGYGQCFNKHALRD 337
> Hs10835059
Length=437
Score = 68.9 bits (167), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 49/168 (29%), Positives = 79/168 (47%), Gaps = 21/168 (12%)
Query 1 SLSIAGQLHRIDVDQLGWWLAERQMA-SGGLNGRPEKLPDLCYSWWVLSSLAIIGRL--- 56
+L I + +++ L W+ RQM GG GR KL D CYS+W L ++ R
Sbjct 258 ALVILKRERSLNLKSLLQWVTSRQMRFEGGFQGRCNKLVDGCYSFWQAGLLPLLHRALHA 317
Query 57 ---------HWINKRKLLQ-FILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLSHDY 106
HW+ ++ LQ +IL C GG D+PG D +HT + ++GLS+ H +
Sbjct 318 QGDPALSMSHWMFHQQALQEYILMCCQCPAGGLLDKPGKSRDFYHTCYCLSGLSIAQH-F 376
Query 107 G-----QDLEVPAAADAASTSMDVADETSDKIDVNKISEFIRKVNPVF 149
G D+ + +A + V + DK+ + + F++K P F
Sbjct 377 GSGAMLHDVVLGVPENALQPTHPVYNIGPDKV-IQATTYFLQKPVPGF 423
Score = 28.5 bits (62), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 30/56 (53%), Gaps = 4/56 (7%)
Query 36 KLPDLCYSWWVLSSLAIIGR---LHWINKRKLLQFILACQDPETGGFADRPGNMVD 88
+ P L ++ +++L IIG IN+ KLLQ++ + + P+ G F G VD
Sbjct 146 QYPHLAPTYAAVNALCIIGTEEAYDIINREKLLQYLYSLKQPD-GSFLMHVGGEVD 200
> 7300143
Length=419
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 37/114 (32%), Positives = 59/114 (51%), Gaps = 11/114 (9%)
Query 2 LSIAGQLHRIDVDQLGWWLAERQMA-SGGLNGRPEKLPDLCYSWWVLSSLAII-GRLHWI 59
L++ + + D L W RQM GG GR KL D CYS+WV +++ I L +
Sbjct 249 LALLNEADKCDRQALLKWTLRRQMTYEGGFQGRTNKLVDGCYSFWVGATIPITQATLSGV 308
Query 60 NKR---------KLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLSH 104
+K+ L ++IL C ++GG D+PG D +HT + ++G+S+ H
Sbjct 309 DKQMEHTLFDVEALQEYILLCCQKQSGGLIDKPGKPQDLYHTCYTLSGVSIAQH 362
Score = 37.4 bits (85), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 35/72 (48%), Gaps = 0/72 (0%)
Query 17 GWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWINKRKLLQFILACQDPET 76
G W+A+ Q GG G P Y++ ++ LA++ +++ LL++ L Q
Sbjct 216 GDWIAQCQTYEGGFGGAPGLEAHGGYTFCGIAGLALLNEADKCDRQALLKWTLRRQMTYE 275
Query 77 GGFADRPGNMVD 88
GGF R +VD
Sbjct 276 GGFQGRTNKLVD 287
Score = 28.5 bits (62), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 17/37 (45%), Positives = 20/37 (54%), Gaps = 1/37 (2%)
Query 68 ILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLSH 104
I CQ E GGF PG +T GIAGL+LL+
Sbjct 219 IAQCQTYE-GGFGGAPGLEAHGGYTFCGIAGLALLNE 254
> CE09598
Length=401
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 36/112 (32%), Positives = 53/112 (47%), Gaps = 11/112 (9%)
Query 1 SLSIAGQLHRIDVDQLGWWLAERQMA-SGGLNGRPEKLPDLCYSWWVLSSLAII-GRLH- 57
SL + + D++ L W RQM GG GR KL D CYS+W + ++ G +
Sbjct 246 SLVLLNRFRLADMEGLLRWATRRQMRFEGGFQGRTNKLVDGCYSFWQGAIFPLLDGEMER 305
Query 58 --------WINKRKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSL 101
R L ++IL GGF D+P VD +HT + ++GLS+
Sbjct 306 EGRSLEKGLFEARMLEEYILVGCQSVHGGFKDKPDKPVDLYHTCYVLSGLSV 357
Score = 33.9 bits (76), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 22/84 (26%), Positives = 36/84 (42%), Gaps = 0/84 (0%)
Query 19 WLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWINKRKLLQFILACQDPETGG 78
W+ Q GG G P Y++ ++SL ++ R + LL++ Q GG
Sbjct 216 WIISCQSFEGGFGGEPYTEAHGGYTFCAVASLVLLNRFRLADMEGLLRWATRRQMRFEGG 275
Query 79 FADRPGNMVDPFHTVFGIAGLSLL 102
F R +VD ++ + A LL
Sbjct 276 FQGRTNKLVDGCYSFWQGAIFPLL 299
> YGL155w
Length=376
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 45/94 (47%), Gaps = 5/94 (5%)
Query 24 QMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHW---INKRKLLQFIL-ACQDPETGGF 79
Q GG GR K D CY++W L+SL ++ + W + ++L Q TGGF
Sbjct 271 QSDDGGFQGRENKFADTCYAFWCLNSLHLLTK-DWKMLCQTELVTNYLLDRTQKTLTGGF 329
Query 80 ADRPGNMVDPFHTVFGIAGLSLLSHDYGQDLEVP 113
+ D +H+ G A L+L+ + +L +P
Sbjct 330 SKNDEEDADLYHSCLGSAALALIEGKFNGELCIP 363
> At5g40280
Length=482
Score = 42.0 bits (97), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 32/60 (53%), Gaps = 0/60 (0%)
Query 1 SLSIAGQLHRIDVDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLHWIN 60
++ + ++ R+++D L W RQ G GR KL D CY++W + ++ RL+ N
Sbjct 266 AMILINEVDRLNLDSLMNWAVHRQGVEMGFQGRTNKLVDGCYTFWQAAPCVLLQRLYSTN 325
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 34/64 (53%), Gaps = 8/64 (12%)
Query 58 WINKRKLL--------QFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLSHDYGQD 109
+IN+R L +++L C GGF D+P D +HT + ++GLS+ H + +D
Sbjct 380 YINRRMQLVFDSLGLQRYVLLCSKIPDGGFRDKPRKPRDFYHTCYCLSGLSVAQHAWLKD 439
Query 110 LEVP 113
+ P
Sbjct 440 EDTP 443
Score = 33.5 bits (75), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 28/112 (25%), Positives = 51/112 (45%), Gaps = 4/112 (3%)
Query 1 SLSIAGQLHRID---VDQLGWWLAERQMASGGLNGRPEKLPDLCYSWWVLSSLAIIGRLH 57
++S+A L+ +D LG ++ Q GG+ G P Y++ L+++ +I +
Sbjct 215 AISVASILNIMDDELTQGLGDYILSCQTYEGGIGGEPGSEAHGGYTYCGLAAMILINEVD 274
Query 58 WINKRKLLQFILACQDPETGGFADRPGNMVDPFHTVFGIAGLSLLSHDYGQD 109
+N L+ + + Q E GF R +VD +T + A LL Y +
Sbjct 275 RLNLDSLMNWAVHRQGVEM-GFQGRTNKLVDGCYTFWQAAPCVLLQRLYSTN 325
> CE01209
Length=1039
Score = 29.6 bits (65), Expect = 3.0, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 21/44 (47%), Gaps = 7/44 (15%)
Query 77 GGFADRPGNMVDPFHTVFGIAGLSLLSH-------DYGQDLEVP 113
GGF D PG V+P+ + G +L SH DY D+ P
Sbjct 860 GGFEDYPGTRVEPYENDWYSHGYTLYSHTRDAAYADYMHDVCKP 903
> At3g60160
Length=1490
Score = 28.1 bits (61), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 15/52 (28%), Positives = 32/52 (61%), Gaps = 2/52 (3%)
Query 100 SLLSHDYGQDLEVPAAADAASTSMDVADETSDKIDVNKISEFIRKVNPVFCM 151
+L+SH Y + L + + + + TS ++ + S +DV +I++FI VN ++ +
Sbjct 388 ALISHIYQKGLVLSSQSRQSHTSGEIINYMS--VDVQRITDFIWYVNNIWML 437
Lambda K H
0.322 0.138 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2454498488
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40