bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1063_orf1
Length=126
Score E
Sequences producing significant alignments: (Bits) Value
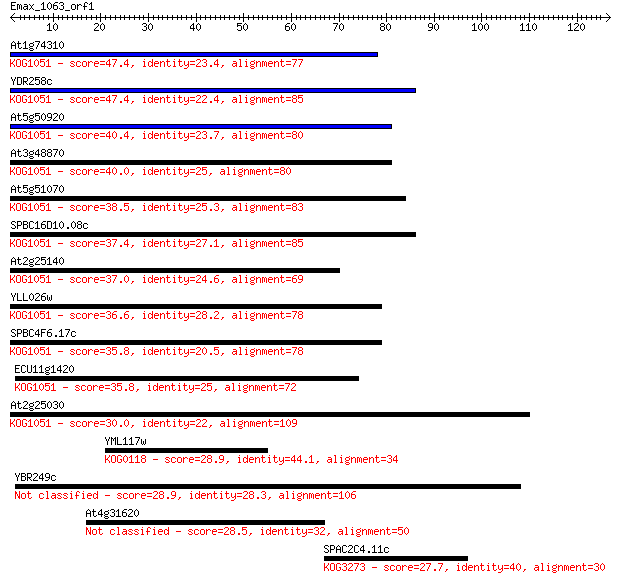
At1g74310 47.4 7e-06
YDR258c 47.4 7e-06
At5g50920 40.4 0.001
At3g48870 40.0 0.001
At5g51070 38.5 0.003
SPBC16D10.08c 37.4 0.007
At2g25140 37.0 0.008
YLL026w 36.6 0.013
SPBC4F6.17c 35.8 0.019
ECU11g1420 35.8 0.022
At2g25030 30.0 1.1
YML117w 28.9 2.5
YBR249c 28.9 2.5
At4g31620 28.5 3.4
SPAC2C4.11c 27.7 4.9
> At1g74310
Length=911
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 18/77 (23%), Positives = 47/77 (61%), Gaps = 0/77 (0%)
Query 1 IFEPLTDNAMRGVLDLMFSHLIKELNKKGIKLEVTNSAKEFILGRAWSHKFGGRRMAKYI 60
+F+PL+ + +R V L + L ++G+ L VT++A ++IL ++ +G R + +++
Sbjct 764 VFDPLSHDQLRKVARLQMKDVAVRLAERGVALAVTDAALDYILAESYDPVYGARPIRRWM 823
Query 61 EKYITAKIAPLLLSAKL 77
EK + +++ +++ ++
Sbjct 824 EKKVVTELSKMVVREEI 840
> YDR258c
Length=811
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 19/85 (22%), Positives = 47/85 (55%), Gaps = 0/85 (0%)
Query 1 IFEPLTDNAMRGVLDLMFSHLIKELNKKGIKLEVTNSAKEFILGRAWSHKFGGRRMAKYI 60
+F L+ +R ++D+ + + L +K +K+++T+ AK+++ + + +G R + + I
Sbjct 702 VFNRLSKKVLRSIVDIRIAEIQDRLAEKRMKIDLTDEAKDWLTDKGYDQLYGARPLNRLI 761
Query 61 EKYITAKIAPLLLSAKLRRGHKAQL 85
+ I +A LL ++R G ++
Sbjct 762 HRQILNSMATFLLKGQIRNGETVRV 786
> At5g50920
Length=929
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/80 (23%), Positives = 44/80 (55%), Gaps = 0/80 (0%)
Query 1 IFEPLTDNAMRGVLDLMFSHLIKELNKKGIKLEVTNSAKEFILGRAWSHKFGGRRMAKYI 60
+F LT ++ + D++ + + L KK I+L+VT KE ++ ++ +G R + + I
Sbjct 813 VFRQLTKLEVKEIADILLKEVFERLKKKEIELQVTERFKERVVDEGYNPSYGARPLRRAI 872
Query 61 EKYITAKIAPLLLSAKLRRG 80
+ + +A +L+ +++ G
Sbjct 873 MRLLEDSMAEKMLAREIKEG 892
> At3g48870
Length=952
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/80 (25%), Positives = 41/80 (51%), Gaps = 0/80 (0%)
Query 1 IFEPLTDNAMRGVLDLMFSHLIKELNKKGIKLEVTNSAKEFILGRAWSHKFGGRRMAKYI 60
+F LT ++ + D+M ++ L K I+L+VT KE ++ + +G R + + I
Sbjct 834 VFRQLTKLEVKEIADIMLKEVVARLEVKEIELQVTERFKERVVDEGFDPSYGARPLRRAI 893
Query 61 EKYITAKIAPLLLSAKLRRG 80
+ + +A +LS ++ G
Sbjct 894 MRLLEDSMAEKMLSRDIKEG 913
> At5g51070
Length=945
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 21/83 (25%), Positives = 37/83 (44%), Gaps = 0/83 (0%)
Query 1 IFEPLTDNAMRGVLDLMFSHLIKELNKKGIKLEVTNSAKEFILGRAWSHKFGGRRMAKYI 60
IF L M +L+LM L L G+ LEV+ KE I + + +G R + + +
Sbjct 833 IFRQLEKAQMMEILNLMLQDLKSRLVALGVGLEVSEPVKELICKQGYDPAYGARPLRRTV 892
Query 61 EKYITAKIAPLLLSAKLRRGHKA 83
+ + ++ L+ + G A
Sbjct 893 TEIVEDPLSEAFLAGSFKPGDTA 915
> SPBC16D10.08c
Length=905
Score = 37.4 bits (85), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 23/87 (26%), Positives = 45/87 (51%), Gaps = 2/87 (2%)
Query 1 IFEPLTDNAMRGVLDLMFSHLIKEL--NKKGIKLEVTNSAKEFILGRAWSHKFGGRRMAK 58
IF L +R +++ + K L N + IK+EV++ AK+ + +S +G R + +
Sbjct 781 IFNRLRRVDIRNIVENRILEVQKRLQSNHRSIKIEVSDEAKDLLGSAGYSPAYGARPLNR 840
Query 59 YIEKYITAKIAPLLLSAKLRRGHKAQL 85
I+ + +A L+L+ +LR A +
Sbjct 841 VIQNQVLNPMAVLILNGQLRDKETAHV 867
> At2g25140
Length=874
Score = 37.0 bits (84), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 17/69 (24%), Positives = 36/69 (52%), Gaps = 0/69 (0%)
Query 1 IFEPLTDNAMRGVLDLMFSHLIKELNKKGIKLEVTNSAKEFILGRAWSHKFGGRRMAKYI 60
+F+PL N + +++L + L +K IKL+ T A + + + +G R + + I
Sbjct 763 VFQPLDSNEISKIVELQMRRVKNSLEQKKIKLQYTKEAVDLLAQLGFDPNYGARPVKRVI 822
Query 61 EKYITAKIA 69
++ + +IA
Sbjct 823 QQMVENEIA 831
> YLL026w
Length=908
Score = 36.6 bits (83), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 22/80 (27%), Positives = 41/80 (51%), Gaps = 2/80 (2%)
Query 1 IFEPLTDNAMRGVLDLMFSHLIK--ELNKKGIKLEVTNSAKEFILGRAWSHKFGGRRMAK 58
IF L+ A+ ++D+ + + E N K KL +T AK+F+ +S G R + +
Sbjct 771 IFNKLSRKAIHKIVDIRLKEIEERFEQNDKHYKLNLTQEAKDFLAKYGYSDDMGARPLNR 830
Query 59 YIEKYITAKIAPLLLSAKLR 78
I+ I K+A +L +++
Sbjct 831 LIQNEILNKLALRILKNEIK 850
> SPBC4F6.17c
Length=803
Score = 35.8 bits (81), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 16/78 (20%), Positives = 44/78 (56%), Gaps = 0/78 (0%)
Query 1 IFEPLTDNAMRGVLDLMFSHLIKELNKKGIKLEVTNSAKEFILGRAWSHKFGGRRMAKYI 60
+F L++ + ++++ + + LN + I L VT +A++++ + +S +G R + + I
Sbjct 696 VFNKLSEKNLEDIVNVRLDEVQQRLNDRRIILTVTEAARKWLAEKGYSPAYGARPLNRLI 755
Query 61 EKYITAKIAPLLLSAKLR 78
+K I +A ++ +++
Sbjct 756 QKRILNTMAMKIIQGEIK 773
> ECU11g1420
Length=851
Score = 35.8 bits (81), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 18/72 (25%), Positives = 37/72 (51%), Gaps = 0/72 (0%)
Query 2 FEPLTDNAMRGVLDLMFSHLIKELNKKGIKLEVTNSAKEFILGRAWSHKFGGRRMAKYIE 61
F L M +L+ +L K+L +GI +V+++ KE ++ + S +G R++ ++I
Sbjct 746 FNNLDRENMHRILEYQMGYLEKKLRGRGITFKVSDAVKEDMVTKVLSSIYGARQLRRFIR 805
Query 62 KYITAKIAPLLL 73
+ +LL
Sbjct 806 SSFKRALTKVLL 817
> At2g25030
Length=265
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 24/109 (22%), Positives = 46/109 (42%), Gaps = 3/109 (2%)
Query 1 IFEPLTDNAMRGVLDLMFSHLIKELNKKGIKLEVTNSAKEFILGRAWSHKFGGRRMAKYI 60
+ +PL + + +++L + K L + I LE T A + + + G R + + I
Sbjct 158 VSQPLNSSEISKIVELQMRQVKKRLEQNKINLEYTKEAVDLLAQLGFDPNNGARPVKQMI 217
Query 61 EKYITAKIAPLLLSAKLRRGHKAQLKRAPNQPNQLNLIICEEDANGECK 109
EK + +I +L + +QPN L+I + + N K
Sbjct 218 EKLVKKEITLKVLKGDF--AEDGTILIDADQPNN-KLVIKKMENNAHVK 263
> YML117w
Length=1134
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 20/34 (58%), Gaps = 3/34 (8%)
Query 21 LIKELNKKGIKLEVTNSAKEFILGRAWSHKFGGR 54
L++E+NK+ V N + E L RA KFGGR
Sbjct 815 LVEEMNKEST---VQNESGEVTLKRATEEKFGGR 845
> YBR249c
Length=370
Score = 28.9 bits (63), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 30/115 (26%), Positives = 47/115 (40%), Gaps = 16/115 (13%)
Query 2 FEPLTDNAMRGVLDL----MFSHLIKELNKKGIKLEVTNSAKE--FILGRAWSHKFGGRR 55
F+ TD + +D SH + K G+ T E F++ R GG++
Sbjct 200 FKNGTDGTLNVAVDACQAAAHSHHFMGVTKHGVAAITTTKGNEHCFVILR------GGKK 253
Query 56 MAKYIEKYITAKIAPLLLSAK---LRRGHKAQLKRAPNQPNQLNLIICEEDANGE 107
Y K + A L + + H K NQP ++N ++CE+ ANGE
Sbjct 254 GTNYDAKSVAEAKAQLPAGSNGLMIDYSHGNSNKDFRNQP-KVNDVVCEQIANGE 307
> At4g31620
Length=1021
Score = 28.5 bits (62), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 23/50 (46%), Gaps = 4/50 (8%)
Query 17 MFSHLIKELNKKGIKLEVTNSAKEFILGRAWSHKFGGRRMAKYIEKYITA 66
FS ++ N + KL++T+ A R W K GRR A E + T
Sbjct 563 FFSTYVEGKNHQSTKLKLTSDA----FDRTWEVKLNGRRFAGGWENFSTV 608
> SPAC2C4.11c
Length=258
Score = 27.7 bits (60), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 67 KIAPLLLSAKLRRGHKAQLKRAPNQPNQLN 96
K AP SA+ +RG K Q++R P P+++
Sbjct 63 KFAPAKTSAEKKRGAKPQMRRVPIPPHRMT 92
Lambda K H
0.320 0.135 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1180352192
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40