bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1027_orf1
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
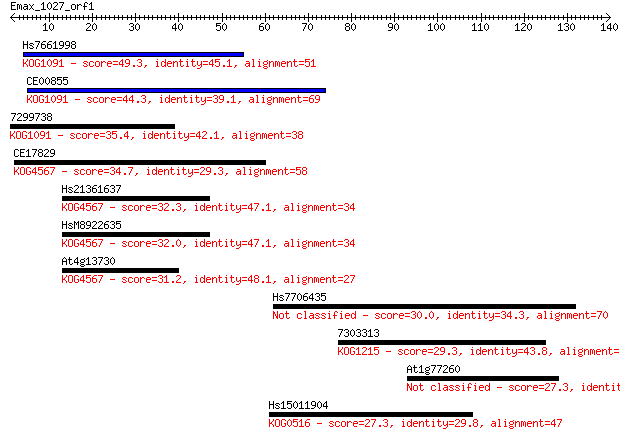
Hs7661998 49.3 2e-06
CE00855 44.3 7e-05
7299738 35.4 0.032
CE17829 34.7 0.060
Hs21361637 32.3 0.31
HsM8922635 32.0 0.39
At4g13730 31.2 0.65
Hs7706435 30.0 1.5
7303313 29.3 2.5
At1g77260 27.3 8.6
Hs15011904 27.3 9.1
> Hs7661998
Length=795
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 23/51 (45%), Positives = 31/51 (60%), Gaps = 0/51 (0%)
Query 4 LPLVDYFAIAMLKFIRRQLLESDYSNCLRRLLKYPPIESVQCLVALSLAVR 54
L LVDY +AML +IR L+ S+Y CL L+ YP I V L+ +L +R
Sbjct 362 LGLVDYIFVAMLLYIRDALISSNYQTCLGLLMHYPFIGDVHSLILKALFLR 412
> CE00855
Length=585
Score = 44.3 bits (103), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 38/81 (46%), Gaps = 12/81 (14%)
Query 5 PLVDYFAIAMLKFIRRQLLESDYSNCLRRLLKYPPIESVQCLVALSLAVR---------- 54
PL +++L IR LL SDY CL+ L++YPPI + V L+ R
Sbjct 326 PLAKCMFVSLLVQIRHLLLSSDYGGCLQYLMRYPPIADIDSFVKLARHYRNPKKNAKPMI 385
Query 55 KGTTLS--TASAAATPNSTNR 73
K S T + ++ PN T R
Sbjct 386 KSNNFSHITVAGSSHPNRTQR 406
> 7299738
Length=654
Score = 35.4 bits (80), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 1 AKKLPLVDYFAIAMLKFIRRQLLESDYSNCLRRLLKYP 38
+ + L +Y +AML IR +LL SDY+ L L++YP
Sbjct 347 SDRFDLPNYILVAMLVHIRDKLLLSDYTTSLTYLMRYP 384
> CE17829
Length=484
Score = 34.7 bits (78), Expect = 0.060, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 31/58 (53%), Gaps = 1/58 (1%)
Query 2 KKLPLVDYFAIAMLKFIRRQLLESDYSNCLRRLLKYPPIESVQCLVALSLAVRKGTTL 59
++ L+ Y +AM++ R L+ D+ C+R L YP + + +VA + +R G L
Sbjct 396 QRFALLQYVCLAMMELKREPLINGDFPFCVRLLQNYPDTD-IAKIVAFAQDIRDGKAL 452
> Hs21361637
Length=400
Score = 32.3 bits (72), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 13 AMLKFIRRQLLESDYSNCLRRLLKYPPIESVQCL 46
AML IR QLLE D++ +R L YP + Q L
Sbjct 357 AMLMLIREQLLEGDFTVNMRLLQDYPITDVCQIL 390
> HsM8922635
Length=275
Score = 32.0 bits (71), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 13 AMLKFIRRQLLESDYSNCLRRLLKYPPIESVQCL 46
AML IR QLLE D++ +R L YP + Q L
Sbjct 232 AMLMLIREQLLEGDFTVNMRLLQDYPITDVCQIL 265
> At4g13730
Length=449
Score = 31.2 bits (69), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 19/27 (70%), Gaps = 0/27 (0%)
Query 13 AMLKFIRRQLLESDYSNCLRRLLKYPP 39
AML +RR+LL D+++ L+ L YPP
Sbjct 407 AMLILVRRRLLAGDFTSNLKLLQNYPP 433
> Hs7706435
Length=543
Score = 30.0 bits (66), Expect = 1.5, Method: Composition-based stats.
Identities = 24/74 (32%), Positives = 31/74 (41%), Gaps = 7/74 (9%)
Query 62 ASAAATPNSTNRRRDNSSNGINNHSSMDRSNSRTANASPAVQGPIRSPPGHPRFIQQPPP 121
A A N+ + S+ + SMDR NS+ A G RSP G P Q PP
Sbjct 219 AEEAKKCRPPNKPQKGPSHDLPRRHSMDRQNSQEKAVGAAAYGA-RSPGGSPG--QSPPT 275
Query 122 ----LTPTIVSNPQ 131
L+P S P+
Sbjct 276 GYSILSPAHFSGPR 289
> 7303313
Length=1612
Score = 29.3 bits (64), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 28/60 (46%), Gaps = 12/60 (20%)
Query 77 NSSNGINNHS--SMDRSNSRTANASPAVQGPIRSPPG----------HPRFIQQPPPLTP 124
NS N +N++ + ++S T N S V PI PP H + I QPPP TP
Sbjct 1454 NSRNSMNSYDRNHITGASSSTTNGSSMVAYPINPPPSPATRSRRPYRHYKIINQPPPPTP 1513
> At1g77260
Length=655
Score = 27.3 bits (59), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 22/42 (52%), Gaps = 7/42 (16%)
Query 93 SRTANASPAVQGPIRS-------PPGHPRFIQQPPPLTPTIV 127
S ++N + A+Q I S PP PR PPPL PT+V
Sbjct 50 SSSSNVTEAIQTNITSVAAVAPSPPPRPRLKISPPPLPPTVV 91
> Hs15011904
Length=5430
Score = 27.3 bits (59), Expect = 9.1, Method: Composition-based stats.
Identities = 14/47 (29%), Positives = 23/47 (48%), Gaps = 0/47 (0%)
Query 61 TASAAATPNSTNRRRDNSSNGINNHSSMDRSNSRTANASPAVQGPIR 107
+A + + +S + NS G+N S + + +T ASP GP R
Sbjct 5384 SACSDTSESSAAGGQGNSRRGLNKPSKIPTMSKKTTTASPRTPGPKR 5430
Lambda K H
0.314 0.128 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1534984332
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40