bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0990_orf1
Length=325
Score E
Sequences producing significant alignments: (Bits) Value
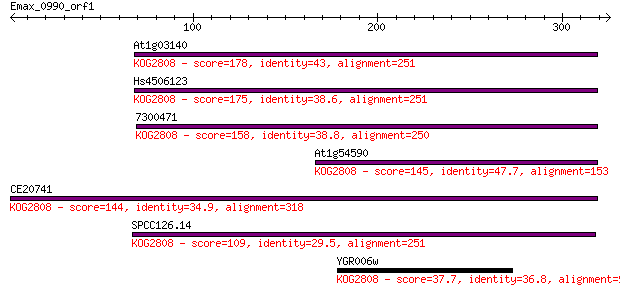
At1g03140 178 1e-44
Hs4506123 175 9e-44
7300471 158 1e-38
At1g54590 145 8e-35
CE20741 144 2e-34
SPCC126.14 109 9e-24
YGR006w 37.7 0.030
> At1g03140
Length=420
Score = 178 bits (452), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 108/272 (39%), Positives = 148/272 (54%), Gaps = 21/272 (7%)
Query 68 QEIFRRLRKLKQPITLFAETPWKRYCRL---CKLEVQVIDDEMTEGQKNVF-------HA 117
QE+ RRLR LKQP+TLF E R RL K + +D +MTEGQ N F
Sbjct 107 QEVIRRLRFLKQPMTLFGEDDQSRLDRLKYVLKEGLFEVDSDMTEGQTNDFLRDIAELKK 166
Query 118 MQREG----------EEEEEFEEESKPVPSSEKGEAEKSDAAAATQNAKSKETTEQ-TKE 166
Q+ G +E E + E E SD A + K E E
Sbjct 167 RQKSGMMGDRKRKSRDERGRDEGDRGETREDELSGGESSDVDADKDMKRLKANFEDLCDE 226
Query 167 AAVIGWARKMLSLWEEELKSRSEDEKATAEGMQATALHRQTKKDLKPLFKKLKHRDLEAD 226
++ + +K+L W++EL + E+ TA+G Q A +Q + L PLF + + L AD
Sbjct 227 DKILVFYKKLLIEWKQELDAMENTERRTAKGKQMVATFKQCARYLVPLFNLCRKKGLPAD 286
Query 227 ILEKLFDIVSLCEERKYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHI 286
I + L +V+ C +R Y A ++ LAIGNA WP+GVTMVGIHER+ R K+ T+ VAHI
Sbjct 287 IRQALMVMVNHCIKRDYLAAMDHYIKLAIGNAPWPIGVTMVGIHERSAREKIYTNSVAHI 346
Query 287 LNDETTRKYIQMFKRLMSFAQRKFPASPSQTI 318
+NDETTRKY+Q KRLM+F QR++P PS+ +
Sbjct 347 MNDETTRKYLQSVKRLMTFCQRRYPTMPSKAV 378
> Hs4506123
Length=342
Score = 175 bits (444), Expect = 9e-44, Method: Compositional matrix adjust.
Identities = 97/262 (37%), Positives = 150/262 (57%), Gaps = 17/262 (6%)
Query 68 QEIFRRLRKLKQPITLFAETPWKRYCRLCKLEVQVIDDEMTEGQKNVFHAMQREGEEEEE 127
QE+ RRLR+ +PI LF ET + + RL K+E+ + E+ +G +N A + +++
Sbjct 82 QEVIRRLRERGEPIRLFGETDYDAFQRLRKIEI--LTPEVNKGLRNDLKAALDKIDQQYL 139
Query 128 FEEESKPVPSSEKGEAEKSDAAAATQNAKSKETTEQTKEA-----------AVIGWARKM 176
E V E GE + + + + E E E+ + + + +
Sbjct 140 NE----IVGGQEPGEEDTQNDLKVHEENTTIEELEALGESLGKGDDHKDMDIITKFLKFL 195
Query 177 LSLWEEELKSRSEDEKATAEGMQATALHRQTKKDLKPLFKKLKHRDLEADILEKLFDIVS 236
L +W +EL +R + K + +G +A +QT+ L+PLF+KL+ R+L ADI E + DI+
Sbjct 196 LGVWAKELNAREDYVKRSVQGKLNSATQKQTESYLRPLFRKLRKRNLPADIKESITDIIK 255
Query 237 LCEERKYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHILNDETTRKYI 296
+R+Y A+ A++ +AIGNA WP+GVTMVGIH R GR K+ + VAH+LNDET RKYI
Sbjct 256 FMLQREYVKANDAYLQMAIGNAPWPIGVTMVGIHARTGREKIFSKHVAHVLNDETQRKYI 315
Query 297 QMFKRLMSFAQRKFPASPSQTI 318
Q KRLM+ Q+ FP PS+ +
Sbjct 316 QGLKRLMTICQKHFPTDPSKCV 337
> 7300471
Length=340
Score = 158 bits (400), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 97/260 (37%), Positives = 142/260 (54%), Gaps = 16/260 (6%)
Query 69 EIFRRLRKLKQPITLFAETPWKRYCRLCKLEVQVIDDEMTEGQKNVFH-AMQREGEEEEE 127
E+ RRLR+ +PI +F ET + + RL + E+ E G +N F AM E +
Sbjct 78 EVIRRLRERGEPILIFGETEPEAFDRLRQCEIS--QPEANRGFRNDFQEAM--EQVDAAY 133
Query 128 FEEESKPVPSSEKGEAEKSDAAAATQN---------AKSKETTEQTKEAAVIGWARKMLS 178
+E P+++ E +KSD A ++ A + + +I +L
Sbjct 134 LQEMFANTPTTK--EDKKSDFAELDESVSWESIQTMAANMGRNKDYDMDVIITLLTFLLK 191
Query 179 LWEEELKSRSEDEKATAEGMQATALHRQTKKDLKPLFKKLKHRDLEADILEKLFDIVSLC 238
LW +++ + S+ EK + + ++ QTK+ +KPLF+KLKH L DIL+ L DI
Sbjct 192 LWNDQIANYSKHEKMSTKVKMTRVIYTQTKEYVKPLFRKLKHHTLPEDILDSLRDICKHL 251
Query 239 EERKYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHILNDETTRKYIQM 298
R Y A A++ +AIGNA WP+GVTMVGIH R GR K+ + VAH++NDET RKYIQ
Sbjct 252 LNRNYITASDAYLEMAIGNAPWPIGVTMVGIHARTGREKIFSKNVAHVMNDETQRKYIQG 311
Query 299 FKRLMSFAQRKFPASPSQTI 318
KRLM+ Q FP PS+ +
Sbjct 312 LKRLMTKCQEYFPTDPSKCV 331
> At1g54590
Length=256
Score = 145 bits (367), Expect = 8e-35, Method: Compositional matrix adjust.
Identities = 73/154 (47%), Positives = 104/154 (67%), Gaps = 1/154 (0%)
Query 166 EAAVIGWARKMLSLWEEELKSRSEDEKATAEGMQATALHRQTKKDLKPLFKKLKHRDLEA 225
E ++ + +K+L W++EL++ E+ TA G Q A Q + L PLF +++ L A
Sbjct 61 EDKILVFCKKLLLEWKQELEAMENTERRTAIGKQMLATFNQCARYLTPLFHLCRNKCLPA 120
Query 226 DILEKLFDIVSLCEERKYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNT-SQVA 284
DI + L +V+ +R Y DA F+ LAIGNA WP+GVTMVGIHER+ R K++T S VA
Sbjct 121 DIRQGLMVMVNCWIKRDYLDATAQFIKLAIGNAPWPIGVTMVGIHERSAREKISTSSSVA 180
Query 285 HILNDETTRKYIQMFKRLMSFAQRKFPASPSQTI 318
HI+N+ETTRKY+Q KRLM+F QR++ A PS++I
Sbjct 181 HIMNNETTRKYLQSVKRLMTFCQRRYSALPSKSI 214
> CE20741
Length=348
Score = 144 bits (363), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 111/333 (33%), Positives = 159/333 (47%), Gaps = 48/333 (14%)
Query 1 QEKLKEKEELADRKRKAELEQLAELQDRIKARPFQKNPEAEAAAAATTECADTTEGTDEE 60
QE ++++E+A +KRK + E L E R K P + E + E
Sbjct 42 QEYERKQQEIASKKRKVDDEILQESSSRTKPAPVENESEID------------------E 83
Query 61 REPPVELQEIFRRLRKLKQPITLFAETPWKRYCRLCKLEVQVIDDEMTEGQKNVFH-AMQ 119
+ P + EI RLR+ PI LF ET RL +LE+ D + EG +N AM+
Sbjct 84 KTP---MSEIQTRLRQRNHPIMLFGETDIDVRKRLHQLELAQPD--LNEGWENELQTAMK 138
Query 120 REGEEEEEFEEESKPVPSSEKGEAEKSDAAAAT-------QNAKSKETTEQTKEA----- 167
G+E + K V A + D A +N + T +
Sbjct 139 VIGKEMD------KAVVEGTADSATRHDIALPQGYEEDNWKNIEHNSTLLSVDDDLKRDC 192
Query 168 -AVIGWARKMLSLWEEELKSRSEDEKATAEGMQATALHRQTKKDLKPLFKKLKHRDLEAD 226
++ R +L+ W ++L R D K TA+ A H+QT LK L ++ + D
Sbjct 193 DIILSICRYILARWAKDLNDRPLDVKKTAQA----AHHKQTMMHLKSLMTSMERYNCNND 248
Query 227 ILEKLFDIVSL-CEERKYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAH 285
I L I L +R Y +A+ A+M +AIGNA WP+GVT GIH+R G +K S +AH
Sbjct 249 IRHHLAKICRLLVIDRNYLEANNAYMEMAIGNAPWPVGVTRSGIHQRPGSAKSYVSNIAH 308
Query 286 ILNDETTRKYIQMFKRLMSFAQRKFPASPSQTI 318
+LNDET RKYIQ FKRLM+ Q FP PS+++
Sbjct 309 VLNDETQRKYIQAFKRLMTKMQEYFPTDPSKSV 341
> SPCC126.14
Length=343
Score = 109 bits (272), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 74/255 (29%), Positives = 125/255 (49%), Gaps = 42/255 (16%)
Query 67 LQEIFRRLRKLKQPITLFAET---PWKRYCRLCKLE-VQVIDDEMTEGQKNVFHAMQREG 122
L EI +LR++K+PI LF E+ +RY L K + ++ I++E+ +G
Sbjct 126 LTEIIAKLREMKEPIRLFGESEEATIQRYYSLLKYKKLEEIENELLT-----------KG 174
Query 123 EEEEEFEEESKPVPSSEKGEAEKSDAAAATQNAKSKETTEQTKEAAVIGWARKMLSLWEE 182
E +FE A T+ SK+ V+ + + + +W+
Sbjct 175 VETIDFEH------------------ATTTKPKVSKQ---------VVAFLQHGIRIWDN 207
Query 183 ELKSRSEDEKATAEGMQATALHRQTKKDLKPLFKKLKHRDLEADILEKLFDIVSLCEERK 242
L S+S + ++E + RQ K+DL L + + L DI + + +I C++ +
Sbjct 208 FLSSKSINSFESSESQMQLKIFRQAKQDLDVLIQLIVDEALNDDIFKSIAEICYRCQKHE 267
Query 243 YRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHILNDETTRKYIQMFKRL 302
+ A+ ++ L IGNA WP+GVTMVGIHER+ +L + ++IL DE RK +Q KR
Sbjct 268 FVKANDMYLRLTIGNAPWPIGVTMVGIHERSAHQRLQANPSSNILKDEKKRKCLQALKRF 327
Query 303 MSFAQRKFPASPSQT 317
++F +R+ P T
Sbjct 328 ITFQERESSNLPEYT 342
> YGR006w
Length=219
Score = 37.7 bits (86), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 53/100 (53%), Gaps = 9/100 (9%)
Query 178 SLWEEELKSRSEDEKATAEGMQATALHRQTKKDLKPLFKKLKHRDLEADILEKLFDIV-S 236
+L+ E+ SR KA+ E L TKK L PL +L+ L D+L L ++
Sbjct 114 NLYIHEILSR---WKASLEAYHP-ELFLDTKKALFPLLLQLRRNQLAPDLLISLATVLYH 169
Query 237 LCEERKYRDAHGAFMLLAIGNAAWPMGVT----MVGIHER 272
L + ++ A ++M L+IGN AWP+GVT M+ +H R
Sbjct 170 LQQPKEINLAVQSYMKLSIGNVAWPIGVTSVAFMLVVHIR 209
Lambda K H
0.312 0.126 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7601516496
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40