bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0975_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
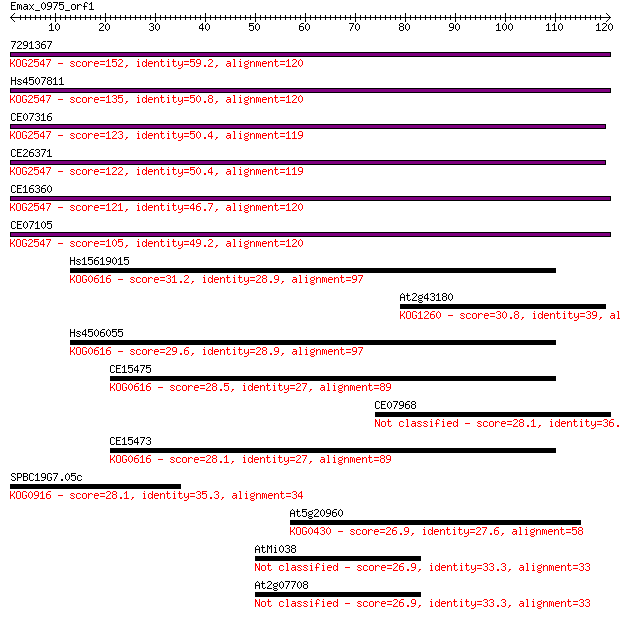
7291367 152 1e-37
Hs4507811 135 1e-32
CE07316 123 1e-28
CE26371 122 1e-28
CE16360 121 3e-28
CE07105 105 2e-23
Hs15619015 31.2 0.47
At2g43180 30.8 0.55
Hs4506055 29.6 1.5
CE15475 28.5 3.3
CE07968 28.1 3.5
CE15473 28.1 4.1
SPBC19G7.05c 28.1 4.6
At5g20960 26.9 8.9
AtMi038 26.9 9.7
At2g07708 26.9 9.7
> 7291367
Length=440
Score = 152 bits (385), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 71/120 (59%), Positives = 94/120 (78%), Gaps = 0/120 (0%)
Query 1 VSDAGILTEENTLLVLVDDMKDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAAD 60
+SD+GI +++TLL +V +M + ALVHQMPF C+R+GF A EKV+FGT + IYL+AD
Sbjct 143 ISDSGIKMKDDTLLDMVQNMSEKHALVHQMPFTCDRDGFAATFEKVFFGTVQSRIYLSAD 202
Query 61 FLRINCLTGMSALMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKIKLSSIPALQNS 120
L INC TGMS L+RK V+D++GG+ AFG YLAEDFF A+S T GWK+++S+ PALQNS
Sbjct 203 VLGINCHTGMSCLLRKAVIDQLGGLRAFGCYLAEDFFIARSVTRLGWKMRISNQPALQNS 262
> Hs4507811
Length=394
Score = 135 bits (341), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 61/120 (50%), Positives = 86/120 (71%), Gaps = 0/120 (0%)
Query 1 VSDAGILTEENTLLVLVDDMKDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAAD 60
+ D+GI +TL +V+ M + V LVH +P+V +R+GF A LE+VYFGT+H Y++A+
Sbjct 142 ICDSGIRVIPDTLTDMVNQMTEKVGLVHGLPYVADRQGFAATLEQVYFGTSHPRYYISAN 201
Query 61 FLRINCLTGMSALMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKIKLSSIPALQNS 120
C+TGMS LMRKDVLD+ GG+ AF QY+AED+F AK+ DRGW+ +S+ A+QNS
Sbjct 202 VTGFKCVTGMSCLMRKDVLDQAGGLIAFAQYIAEDYFMAKAIADRGWRFAMSTQVAMQNS 261
> CE07316
Length=511
Score = 123 bits (308), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 60/121 (49%), Positives = 82/121 (67%), Gaps = 2/121 (1%)
Query 1 VSDAGILTEENTLLVLVDDM--KDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLA 58
VSD+GI + +L + M + +ALV Q P+ +REGF A E++YFGT+H IYLA
Sbjct 206 VSDSGIFMRSDGVLDMATTMMSHEKMALVTQTPYCKDREGFDAAFEQMYFGTSHGRIYLA 265
Query 59 ADFLRINCLTGMSALMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKIKLSSIPALQ 118
+ + C TGMS++M+K+ LDE GGIS FG YLAED+FF + +RG+K +SS PALQ
Sbjct 266 GNCMDFVCSTGMSSMMKKEALDECGGISNFGGYLAEDYFFGRELANRGYKSAISSHPALQ 325
Query 119 N 119
N
Sbjct 326 N 326
> CE26371
Length=522
Score = 122 bits (307), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 60/121 (49%), Positives = 82/121 (67%), Gaps = 2/121 (1%)
Query 1 VSDAGILTEENTLLVLVDDM--KDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLA 58
VSD+GI + +L + M + +ALV Q P+ +REGF A E++YFGT+H IYLA
Sbjct 217 VSDSGIFMRSDGVLDMATTMMSHEKMALVTQTPYCKDREGFDAAFEQMYFGTSHGRIYLA 276
Query 59 ADFLRINCLTGMSALMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKIKLSSIPALQ 118
+ + C TGMS++M+K+ LDE GGIS FG YLAED+FF + +RG+K +SS PALQ
Sbjct 277 GNCMDFVCSTGMSSMMKKEALDECGGISNFGGYLAEDYFFGRELANRGYKSAISSHPALQ 336
Query 119 N 119
N
Sbjct 337 N 337
> CE16360
Length=422
Score = 121 bits (304), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 56/122 (45%), Positives = 84/122 (68%), Gaps = 2/122 (1%)
Query 1 VSDAGILTEENTLLVLVDDM--KDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLA 58
+SD+ I + +L + M + +A V Q+P+ +R+GF A E+++FGT+HA +YL
Sbjct 198 ISDSAIFMRPDGILDMATTMMSHEKMASVTQIPYCKDRQGFHAAFEQIFFGTSHARLYLV 257
Query 59 ADFLRINCLTGMSALMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKIKLSSIPALQ 118
+FL + C +GMS++M+K LDE GG+ FG+YLAED+FFAK+ T RG K +S+ PALQ
Sbjct 258 GNFLGVVCSSGMSSMMKKSALDECGGMEKFGEYLAEDYFFAKALTSRGCKAAISTHPALQ 317
Query 119 NS 120
NS
Sbjct 318 NS 319
> CE07105
Length=420
Score = 105 bits (262), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 59/154 (38%), Positives = 86/154 (55%), Gaps = 34/154 (22%)
Query 1 VSDAGILTEENTLLVLVDDM--KDGVALVHQMPFVCNREGFTA------ILEK------- 45
+SD+GI + + +L + M + +ALV Q P+ +R+G + IL++
Sbjct 189 ISDSGIFMKSDAVLDMASTMMSHETMALVTQTPYCKDRKGKSKNCAPIQILKRLTLIFIV 248
Query 46 -------------------VYFGTAHAWIYLAADFLRINCLTGMSALMRKDVLDEVGGIS 86
+YFGT+HA IYLA + L+ NC TGMS++M+K+ LDE GG +
Sbjct 249 VNVLACFNIALSISVWATLIYFGTSHARIYLAGNCLQFNCPTGMSSMMKKEALDECGGFA 308
Query 87 AFGQYLAEDFFFAKSFTDRGWKIKLSSIPALQNS 120
AF YLAED+FF K RG+K +S+ PALQNS
Sbjct 309 AFSGYLAEDYFFGKKLASRGYKSGISTHPALQNS 342
> Hs15619015
Length=351
Score = 31.2 bits (69), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 38/97 (39%), Gaps = 1/97 (1%)
Query 13 LLVLVDDMKDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAADFLRINCLTGMSA 72
L+ L KD L M +V E F+ + F HA Y A L + L +
Sbjct 104 LVKLQFSFKDNSYLYLVMEYVPGGEMFSRLQRVGRFSEPHACFYAAQVVLAVQYLHSLD- 162
Query 73 LMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKI 109
L+ +D+ E I G DF FAK R W +
Sbjct 163 LIHRDLKPENLLIDQQGYLQVTDFGFAKRVKGRTWTL 199
> At2g43180
Length=492
Score = 30.8 bits (68), Expect = 0.55, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 79 LDEVGGISAFGQYLAEDFFFAKSFTDRGWKIKLSSIPALQN 119
L+E+G I F Y E+ +A S +DRG K S + +L N
Sbjct 330 LEEIGEILGFDTYEEEEKRYATSSSDRGETWKFSVVTSLGN 370
> Hs4506055
Length=351
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 37/97 (38%), Gaps = 1/97 (1%)
Query 13 LLVLVDDMKDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAADFLRINCLTGMSA 72
L+ L KD L M +V E F+ + F HA Y A L L +
Sbjct 104 LVKLEFSFKDNSNLYMVMEYVPGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLD- 162
Query 73 LMRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKI 109
L+ +D+ E I G DF FAK R W +
Sbjct 163 LIYRDLKPENLLIDQQGYIQVTDFGFAKRVKGRTWTL 199
> CE15475
Length=375
Score = 28.5 bits (62), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 35/89 (39%), Gaps = 1/89 (1%)
Query 21 KDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAADFLRINCLTGMSALMRKDVLD 80
KD L + F+ E F+ + F H+ Y A L L + L+ +D+
Sbjct 120 KDNSNLYMVLEFISGGEMFSHLRRIGRFSEPHSRFYAAQIVLAFEYLHSLD-LIYRDLKP 178
Query 81 EVGGISAFGQYLAEDFFFAKSFTDRGWKI 109
E I + G DF FAK R W +
Sbjct 179 ENLLIDSTGYLKITDFGFAKRVKGRTWTL 207
> CE07968
Length=988
Score = 28.1 bits (61), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 26/47 (55%), Gaps = 2/47 (4%)
Query 74 MRKDVLDEVGGISAFGQYLAEDFFFAKSFTDRGWKIKLSSIPALQNS 120
+RK V+D V I+ F Y E K+F +G+K+ L SIP + +
Sbjct 909 VRKIVIDNV--IAMFSIYNNEQSKRMKNFYPKGFKLDLCSIPLMMTA 953
> CE15473
Length=359
Score = 28.1 bits (61), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 34/89 (38%), Gaps = 1/89 (1%)
Query 21 KDGVALVHQMPFVCNREGFTAILEKVYFGTAHAWIYLAADFLRINCLTGMSALMRKDVLD 80
KD L + F+ E F+ + F H+ Y A L L + + R D+
Sbjct 120 KDNSNLYMVLEFISGGEMFSHLRRIGRFSEPHSRFYAAQIVLAFEYLHSLDLIYR-DLKP 178
Query 81 EVGGISAFGQYLAEDFFFAKSFTDRGWKI 109
E I + G DF FAK R W +
Sbjct 179 ENLLIDSTGYLKITDFGFAKRVKGRTWTL 207
> SPBC19G7.05c
Length=1729
Score = 28.1 bits (61), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 22/35 (62%), Gaps = 1/35 (2%)
Query 1 VSDAGILTEEN-TLLVLVDDMKDGVALVHQMPFVC 34
V D +L +EN +++ +D+ K+GV + +PF C
Sbjct 772 VDDTKLLADENDSVIGSIDNEKNGVNKAYDLPFYC 806
> At5g20960
Length=1368
Score = 26.9 bits (58), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 16/76 (21%), Positives = 32/76 (42%), Gaps = 18/76 (23%)
Query 57 LAADFLRINCLTGMSALMRKDVLDEVGG---------------ISAFGQYLAEDFFFAKS 101
+AA + +N LTG + ++R D++ + G + G ++ E+F
Sbjct 1192 IAASEVEVNVLTGETTILRTDIIYDCGKSLNPAVDLGQIEGAFVQGLGFFMLEEFLMNSD 1251
Query 102 ---FTDRGWKIKLSSI 114
TD W K+ ++
Sbjct 1252 GLVVTDSTWTYKIPTV 1267
> AtMi038
Length=141
Score = 26.9 bits (58), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 11/33 (33%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 50 TAHAWIYLAADFLRINCLTGMSALMRKDVLDEV 82
T H W+ L ++ R NC+ +SA + +V +V
Sbjct 37 TMHGWMKLYLEYQRWNCMLRLSASIELEVFMDV 69
> At2g07708
Length=141
Score = 26.9 bits (58), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 11/33 (33%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 50 TAHAWIYLAADFLRINCLTGMSALMRKDVLDEV 82
T H W+ L ++ R NC+ +SA + +V +V
Sbjct 37 TMHGWMKLYLEYQRWNCMLRLSASIELEVFMDV 69
Lambda K H
0.325 0.139 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160968786
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40