bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0939_orf2
Length=126
Score E
Sequences producing significant alignments: (Bits) Value
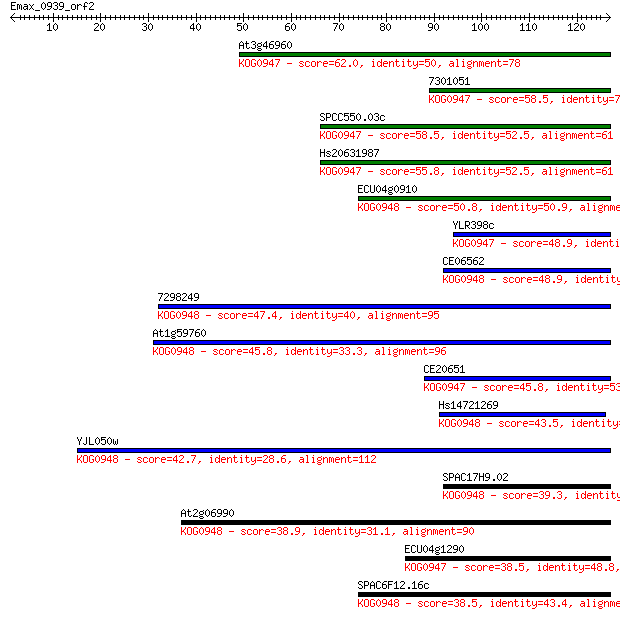
At3g46960 62.0 3e-10
7301051 58.5 3e-09
SPCC550.03c 58.5 3e-09
Hs20631987 55.8 2e-08
ECU04g0910 50.8 5e-07
YLR398c 48.9 2e-06
CE06562 48.9 2e-06
7298249 47.4 7e-06
At1g59760 45.8 2e-05
CE20651 45.8 2e-05
Hs14721269 43.5 9e-05
YJL050w 42.7 2e-04
SPAC17H9.02 39.3 0.002
At2g06990 38.9 0.003
ECU04g1290 38.5 0.003
SPAC6F12.16c 38.5 0.003
> At3g46960
Length=1347
Score = 62.0 bits (149), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 39/86 (45%), Positives = 50/86 (58%), Gaps = 11/86 (12%)
Query 49 AAAAAAAAAAAVTGAA--------AAAPTDTASPSTAAADLSPEEEPLAIEFGFPLDDFQ 100
+A A + AVTG++ A D+ + +L P+ +AIEF F LD+FQ
Sbjct 300 SAKTAIMSEEAVTGSSDKQLRKEGWATKGDSQDIADRFYELVPD---MAIEFPFELDNFQ 356
Query 101 KRAILRLEKNQCVFVAAHTSAGKTVV 126
K AI LEK + VFVAAHTSAGKTVV
Sbjct 357 KEAICCLEKGESVFVAAHTSAGKTVV 382
> 7301051
Length=474
Score = 58.5 bits (140), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 28/38 (73%), Positives = 33/38 (86%), Gaps = 0/38 (0%)
Query 89 AIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
A++F F LD FQK+AIL+LE+ Q VFVAAHTSAGKTVV
Sbjct 256 AMDFPFELDVFQKQAILKLEQRQYVFVAAHTSAGKTVV 293
> SPCC550.03c
Length=1213
Score = 58.5 bits (140), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 32/61 (52%), Positives = 39/61 (63%), Gaps = 3/61 (4%)
Query 66 AAPTDTASPSTAAADLSPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTV 125
A D+++P L PE +A++F F LD+FQK AI LE VFVAAHTSAGKTV
Sbjct 254 AHVVDSSAPIENFQQLVPE---MALDFPFELDNFQKEAIYHLEMGDSVFVAAHTSAGKTV 310
Query 126 V 126
V
Sbjct 311 V 311
> Hs20631987
Length=1246
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 32/61 (52%), Positives = 38/61 (62%), Gaps = 3/61 (4%)
Query 66 AAPTDTASPSTAAADLSPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTV 125
A P D SP L P+ A ++ F D FQK+AIL LE++ VFVAAHTSAGKTV
Sbjct 284 AIPVDATSPVGDFYRLIPQP---AFQWAFEPDVFQKQAILHLERHDSVFVAAHTSAGKTV 340
Query 126 V 126
V
Sbjct 341 V 341
> ECU04g0910
Length=933
Score = 50.8 bits (120), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 27/55 (49%), Positives = 38/55 (69%), Gaps = 2/55 (3%)
Query 74 PSTAAADLSPEE--EPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
P AA L P++ +P A + F LDDFQK ++ LE+++ V V+AHTS+GKTVV
Sbjct 47 PVGAAYTLLPKQFCKPAAKNYLFELDDFQKISVCSLERDESVLVSAHTSSGKTVV 101
> YLR398c
Length=1287
Score = 48.9 bits (115), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 23/33 (69%), Positives = 25/33 (75%), Gaps = 0/33 (0%)
Query 94 FPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
F LD FQK A+ LE+ VFVAAHTSAGKTVV
Sbjct 328 FELDTFQKEAVYHLEQGDSVFVAAHTSAGKTVV 360
> CE06562
Length=1026
Score = 48.9 bits (115), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 23/35 (65%), Positives = 28/35 (80%), Gaps = 0/35 (0%)
Query 92 FGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
+ F LD FQK+AIL ++ NQ V V+AHTSAGKTVV
Sbjct 122 YPFQLDAFQKQAILCIDNNQSVLVSAHTSAGKTVV 156
> 7298249
Length=1055
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 38/95 (40%), Positives = 48/95 (50%), Gaps = 13/95 (13%)
Query 32 SAALESALRAQVAPPPDAAAAAAAAAAAVTGAAAAAPTDTASPSTAAADLSPEEEPLAIE 91
SA LE ALR ++ A + T AA P P L P A E
Sbjct 101 SATLE-ALRTRIV------THLLDAPKSCTHEVAAHPDQEYIP------LKPFSGVPAKE 147
Query 92 FGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
+ F LD FQ++AIL ++ +Q V V+AHTSAGKTVV
Sbjct 148 YPFVLDPFQRQAILCIDNSQSVLVSAHTSAGKTVV 182
> At1g59760
Length=988
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 32/98 (32%), Positives = 44/98 (44%), Gaps = 3/98 (3%)
Query 31 GSAALESALRAQVAPPPDAAAAAAAAAAAVTGAAAAAPTDTASPSTAAADLSP--EEEPL 88
GS +S + + PP + + D + P L+P +P
Sbjct 2 GSVKRKSVEESSDSAPPQKVQREDDSTQIINEELVGCVHDVSFPENYVP-LAPSVHNKPP 60
Query 89 AIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
A +F F LD FQ AI L+ + V V+AHTSAGKTVV
Sbjct 61 AKDFPFTLDSFQSEAIKCLDNGESVMVSAHTSAGKTVV 98
> CE20651
Length=1260
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 21/39 (53%), Positives = 31/39 (79%), Gaps = 0/39 (0%)
Query 88 LAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
+A ++ F LD FQ+ ++L +E+ + +FVAAHTSAGKTVV
Sbjct 279 MARKYPFSLDPFQQSSVLCMERGESLFVAAHTSAGKTVV 317
> Hs14721269
Length=1042
Score = 43.5 bits (101), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 21/35 (60%), Positives = 26/35 (74%), Gaps = 0/35 (0%)
Query 91 EFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTV 125
E+ F LD FQ+ AI ++ NQ V V+AHTSAGKTV
Sbjct 135 EYPFILDAFQREAIQCVDNNQSVLVSAHTSAGKTV 169
> YJL050w
Length=1073
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 32/112 (28%), Positives = 42/112 (37%), Gaps = 26/112 (23%)
Query 15 AAAAAARQQQQQAAAGGSAALESALRAQVAPPPDAAAAAAAAAAAVTGAAAAAPTDTASP 74
A+ + Q G L +R QVA PP+ A V A
Sbjct 95 ASKGLTNSETLQVEQDGKVRLSHQVRHQVALPPNYDYTPIAEHKRVNEART--------- 145
Query 75 STAAADLSPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
+ F LD FQ AI +++ + V V+AHTSAGKTVV
Sbjct 146 -----------------YPFTLDPFQDTAISCIDRGESVLVSAHTSAGKTVV 180
> SPAC17H9.02
Length=1030
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/35 (54%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 92 FGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
+ F LD FQ AI +E+ + V V+AHTSAGKTV+
Sbjct 122 YPFELDPFQSTAIKCVERMESVLVSAHTSAGKTVI 156
> At2g06990
Length=996
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 28/90 (31%), Positives = 41/90 (45%), Gaps = 5/90 (5%)
Query 37 SALRAQVAPPPDAAAAAAAAAAAVTGAAAAAPTDTASPSTAAADLSPEEEPLAIEFGFPL 96
S LR+ P P+ + A A P D +P+ + P + F L
Sbjct 16 SKLRSDETPTPEPRTKRRSLKRACV-HEVAVPND-YTPTKEETIHGTLDNPT---YPFKL 70
Query 97 DDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
D FQ ++ LE+ + + V+AHTSAGKT V
Sbjct 71 DPFQSVSVACLERKESILVSAHTSAGKTAV 100
> ECU04g1290
Length=881
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 21/43 (48%), Positives = 29/43 (67%), Gaps = 2/43 (4%)
Query 84 EEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
+E+ L I GF D FQ++A L + VFV+AHTS+GKT+V
Sbjct 55 DEKILNI--GFEPDTFQRQAFYLLSRASSVFVSAHTSSGKTLV 95
> SPAC6F12.16c
Length=1117
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 23/55 (41%), Positives = 33/55 (60%), Gaps = 2/55 (3%)
Query 74 PSTAAADLSPEEEPL--AIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
P+ +S + P+ A + F LD FQ +I +E+ + V V+AHTSAGKTVV
Sbjct 175 PNYDYVPISKHKSPIPPARTYPFTLDPFQAVSIACIERQESVLVSAHTSAGKTVV 229
Lambda K H
0.314 0.121 0.323
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1180352192
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40