bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0891_orf1
Length=185
Score E
Sequences producing significant alignments: (Bits) Value
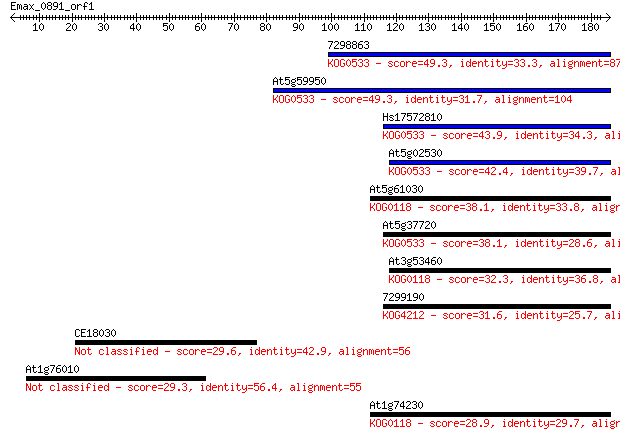
7298863 49.3 5e-06
At5g59950 49.3 5e-06
Hs17572810 43.9 2e-04
At5g02530 42.4 6e-04
At5g61030 38.1 0.009
At5g37720 38.1 0.010
At3g53460 32.3 0.52
7299190 31.6 1.0
CE18030 29.6 3.7
At1g76010 29.3 4.8
At1g74230 28.9 5.6
> 7298863
Length=266
Score = 49.3 bits (116), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 29/96 (30%), Positives = 52/96 (54%), Gaps = 10/96 (10%)
Query 99 VHWSWS----NGPR-----GGSSTAIVRISGLDYSVLDSELRELMEKSCEGILKVWIDYD 149
V+ +W +GP+ GGS + + LDY V +++++EL I K + YD
Sbjct 85 VNSAWKHDMYDGPKRGAVGGGSGPTRLIVGNLDYGVSNTDIKELFNDFGP-IKKAAVHYD 143
Query 150 KTDRSRGTGGCVFRTLADAQRAVETFDGRRIEGKPL 185
++ RS GT +F ADA +A++ + G ++G+P+
Sbjct 144 RSGRSLGTADVIFERRADALKAIKQYHGVPLDGRPM 179
> At5g59950
Length=244
Score = 49.3 bits (116), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 60/113 (53%), Gaps = 10/113 (8%)
Query 82 SNSSSSYGGGGGPNSSGVHWSWSN---GPRGGSSTAIVR------ISGLDYSVLDSELRE 132
S S+ Y P S+ H +S+ R G S+A + IS LDY V++ +++E
Sbjct 47 STRSAPYQSAKAPESTWGHDMFSDRSEDHRSGRSSAGIETGTKLYISNLDYGVMNEDIKE 106
Query 133 LMEKSCEGILKVWIDYDKTDRSRGTGGCVFRTLADAQRAVETFDGRRIEGKPL 185
L + E + + + +D++ RS+GT V+ DA AV+ ++ +++GKP+
Sbjct 107 LFAEVGE-LKRYTVHFDRSGRSKGTAEVVYSRRGDALAAVKKYNDVQLDGKPM 158
> Hs17572810
Length=255
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 44/71 (61%), Gaps = 3/71 (4%)
Query 116 VRISGLDYSVLDSELRELMEKSCEGILK-VWIDYDKTDRSRGTGGCVFRTLADAQRAVET 174
+ +S LD+ V D++++EL + G LK + YD++ RS GT F ADA +A++
Sbjct 106 LLVSNLDFGVSDADIQELFAEF--GTLKKAAVHYDRSGRSLGTANVHFERKADALKAMKQ 163
Query 175 FDGRRIEGKPL 185
++G ++G+P+
Sbjct 164 YNGFPLDGRPM 174
> At5g02530
Length=290
Score = 42.4 bits (98), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 27/69 (39%), Positives = 42/69 (60%), Gaps = 3/69 (4%)
Query 118 ISGLDYSVLDSELRELMEKSCEGILKVW-IDYDKTDRSRGTGGCVFRTLADAQRAVETFD 176
IS LDY V + +++EL S G LK + I YD++ RS+GT VF DA AV+ ++
Sbjct 112 ISNLDYGVSNEDIKELF--SEVGDLKRYGIHYDRSGRSKGTAEVVFSRRGDALAAVKRYN 169
Query 177 GRRIEGKPL 185
+++GK +
Sbjct 170 NVQLDGKLM 178
> At5g61030
Length=309
Score = 38.1 bits (87), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 41/75 (54%), Gaps = 2/75 (2%)
Query 112 STAIVRISGLDYSVLDSELRELMEKSCEGI-LKVWIDYDKTDRSRGTGGCVFRTLADAQR 170
S++ + I G+ YS+ + LRE K E + +V +D + T RSRG G F + A
Sbjct 38 SSSKLFIGGMAYSMDEDSLREAFTKYGEVVDTRVILDRE-TGRSRGFGFVTFTSSEAASS 96
Query 171 AVETFDGRRIEGKPL 185
A++ DGR + G+ +
Sbjct 97 AIQALDGRDLHGRVV 111
> At5g37720
Length=292
Score = 38.1 bits (87), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 40/70 (57%), Gaps = 1/70 (1%)
Query 116 VRISGLDYSVLDSELRELMEKSCEGILKVWIDYDKTDRSRGTGGCVFRTLADAQRAVETF 175
+ ++ LD V + ++REL + E + + I YDK R GT V+ +DA +A++ +
Sbjct 99 LHVTNLDQGVTNEDIRELFSEIGE-VERYAIHYDKNGRPSGTAEVVYPRRSDAFQALKKY 157
Query 176 DGRRIEGKPL 185
+ ++G+P+
Sbjct 158 NNVLLDGRPM 167
> At3g53460
Length=342
Score = 32.3 bits (72), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 38/69 (55%), Gaps = 2/69 (2%)
Query 118 ISGLDYSVLDSELRELMEKSCEGILKVWIDYDK-TDRSRGTGGCVFRTLADAQRAVETFD 176
+ L ++V ++L +L E S + V + YDK T RSRG G T A+ + A + F+
Sbjct 103 VGNLSFNVDSAQLAQLFE-SAGNVEMVEVIYDKVTGRSRGFGFVTMSTAAEVEAAAQQFN 161
Query 177 GRRIEGKPL 185
G EG+PL
Sbjct 162 GYEFEGRPL 170
> 7299190
Length=632
Score = 31.6 bits (70), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 18/70 (25%), Positives = 36/70 (51%), Gaps = 0/70 (0%)
Query 116 VRISGLDYSVLDSELRELMEKSCEGILKVWIDYDKTDRSRGTGGCVFRTLADAQRAVETF 175
V IS + Y +L++L + I V + +D++ ++RG G F+ + Q+A+E
Sbjct 59 VYISNIPYDYRWQDLKDLFRRIVGSIEYVQLFFDESGKARGCGIVEFKDPENVQKALEKM 118
Query 176 DGRRIEGKPL 185
+ + G+ L
Sbjct 119 NRYEVNGREL 128
> CE18030
Length=798
Score = 29.6 bits (65), Expect = 3.7, Method: Composition-based stats.
Identities = 24/65 (36%), Positives = 28/65 (43%), Gaps = 9/65 (13%)
Query 21 GGYRGGRGGG--WNGPYRSYGREG-------TWGPTPGRTRGGGYGGPSPYGRSGPYSTG 71
G +RGG GG W G R G + G RGGG GG +P GR G +
Sbjct 110 GNFRGGDRGGSPWRGGDRGVANRGRGDFSGGSRGGNKFSPRGGGRGGFTPRGRGGNDFSP 169
Query 72 GGGGG 76
GG G
Sbjct 170 RGGRG 174
> At1g76010
Length=350
Score = 29.3 bits (64), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 31/58 (53%), Positives = 35/58 (60%), Gaps = 9/58 (15%)
Query 6 DGPIRRGGYMQERGEGGYRG---GRGGGWNGPYRSYGREGTWGPTPGRTRGGGYGGPS 60
DGP RGGY +G GY G GR GG++GP S GR G GP+ GR GGY GPS
Sbjct 251 DGPQGRGGYDGPQGRRGYDGPPQGR-GGYDGP--SQGRGGYDGPSQGR---GGYDGPS 302
> At1g74230
Length=289
Score = 28.9 bits (63), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 40/75 (53%), Gaps = 3/75 (4%)
Query 112 STAIVRISGLDYSVLDSELRELMEKSCEGI-LKVWIDYDKTDRSRGTGGCVFRTLADAQR 170
S++ + + G+ YS + LRE K E + K+ +D +T RSRG F + +A
Sbjct 32 SSSKIFVGGISYSTDEFGLREAFSKYGEVVDAKIIVDR-ETGRSRGFAFVTFTSTEEASN 90
Query 171 AVETFDGRRIEGKPL 185
A++ DG+ + G+ +
Sbjct 91 AMQ-LDGQDLHGRRI 104
Lambda K H
0.313 0.140 0.454
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2986559618
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40