bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0870_orf1
Length=269
Score E
Sequences producing significant alignments: (Bits) Value
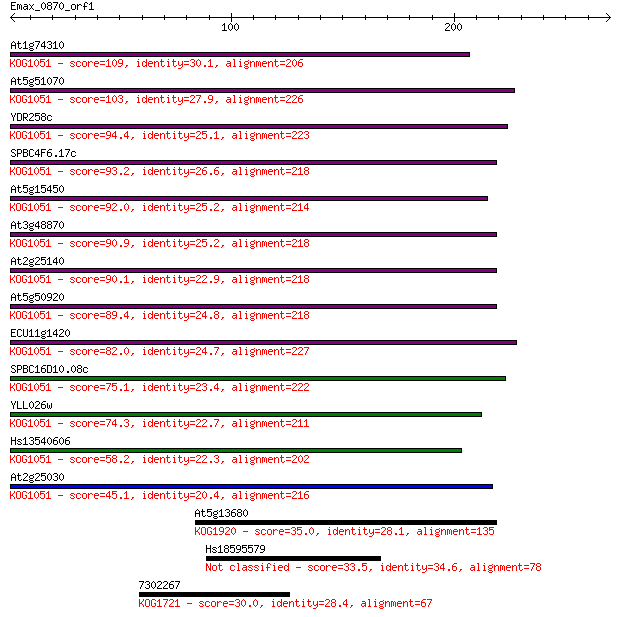
At1g74310 109 8e-24
At5g51070 103 5e-22
YDR258c 94.4 2e-19
SPBC4F6.17c 93.2 6e-19
At5g15450 92.0 1e-18
At3g48870 90.9 3e-18
At2g25140 90.1 5e-18
At5g50920 89.4 7e-18
ECU11g1420 82.0 1e-15
SPBC16D10.08c 75.1 2e-13
YLL026w 74.3 2e-13
Hs13540606 58.2 2e-08
At2g25030 45.1 2e-04
At5g13680 35.0 0.17
Hs18595579 33.5 0.42
7302267 30.0 5.3
> At1g74310
Length=911
Score = 109 bits (272), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 62/206 (30%), Positives = 104/206 (50%), Gaps = 46/206 (22%)
Query 1 FDEVENAHKNLWSLLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQFILEGYKEARAL 60
FDEVE AH +++ LL +LD+G L+D +G VDF N +II+TSN+G + +L G
Sbjct 677 FDEVEKAHVAVFNTLLQVLDDGRLTDGQGRTVDFRNSVIIMTSNLGAEHLLAGL------ 730
Query 61 SGSNEKGGAADAAATGGAEDGSSGKGSGASSVGGFKGTGKSPKDVLRRMRQKVLKEVFGY 120
TGK +V R V++EV +
Sbjct 731 -------------------------------------TGKVTMEV---ARDCVMREVRKH 750
Query 121 FKPQVIGRMSEIIIFEPLNDQALKGVLGLKLSSLTKGLAARGVDFKVADSAMTYILERAA 180
F+P+++ R+ EI++F+PL+ L+ V L++ + LA RGV V D+A+ YIL +
Sbjct 751 FRPELLNRLDEIVVFDPLSHDQLRKVARLQMKDVAVRLAERGVALAVTDAALDYILAESY 810
Query 181 SYKYGGRRMGKYLEKYIKPRVAPLLI 206
YG R + +++EK + ++ +++
Sbjct 811 DPVYGARPIRRWMEKKVVTELSKMVV 836
> At5g51070
Length=945
Score = 103 bits (256), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 63/227 (27%), Positives = 112/227 (49%), Gaps = 36/227 (15%)
Query 1 FDEVENAHKNLWSLLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQFILEGYKEARAL 60
FDE+E AH +++++LL + ++GHL+DS+G RV F N LII+TSN+G I +G +
Sbjct 735 FDEIEKAHPDIFNILLQLFEDGHLTDSQGRRVSFKNALIIMTSNVGSLAIAKGRHGSIGF 794
Query 61 SGSNEKGGAADAAATGGAEDGSSGKGSGASSVGGFKGTGKSPKDVLRRMRQKVLKEVFGY 120
++ E+ +S G M+ V++E+ Y
Sbjct 795 ILDDD-------------EEAASYTG----------------------MKALVVEELKNY 819
Query 121 FKPQVIGRMSEIIIFEPLNDQALKGVLGLKLSSLTKGLAARGVDFKVADSAMTYILERAA 180
F+P+++ R+ EI+IF L + +L L L L L A GV +V++ I ++
Sbjct 820 FRPELLNRIDEIVIFRQLEKAQMMEILNLMLQDLKSRLVALGVGLEVSEPVKELICKQGY 879
Query 181 SYKYGGRRMGKYLEKYIKPRVAPLLISGKLKRGDRAVLARSNT-NPN 226
YG R + + + + ++ ++ ++G K GD A + +T NP+
Sbjct 880 DPAYGARPLRRTVTEIVEDPLSEAFLAGSFKPGDTAFVVLDDTGNPS 926
> YDR258c
Length=811
Score = 94.4 bits (233), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 56/223 (25%), Positives = 105/223 (47%), Gaps = 43/223 (19%)
Query 1 FDEVENAHKNLWSLLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQFILEGYKEARAL 60
FDE E AH ++ LLL +LDEG L+DS G+ VDF N +I++TSNIGQ +L K
Sbjct 612 FDEFEKAHPDVSKLLLQVLDEGKLTDSLGHHVDFRNTIIVMTSNIGQDILLNDTKL---- 667
Query 61 SGSNEKGGAADAAATGGAEDGSSGKGSGASSVGGFKGTGKSPKDVLRRMRQKVLKEVFGY 120
G GK A+ + KV++ +
Sbjct 668 --------------------GDDGKIDTAT-------------------KNKVIEAMKRS 688
Query 121 FKPQVIGRMSEIIIFEPLNDQALKGVLGLKLSSLTKGLAARGVDFKVADSAMTYILERAA 180
+ P+ I R+ +I++F L+ + L+ ++ ++++ + LA + + + D A ++ ++
Sbjct 689 YPPEFINRIDDILVFNRLSKKVLRSIVDIRIAEIQDRLAEKRMKIDLTDEAKDWLTDKGY 748
Query 181 SYKYGGRRMGKYLEKYIKPRVAPLLISGKLKRGDRAVLARSNT 223
YG R + + + + I +A L+ G+++ G+ + +T
Sbjct 749 DQLYGARPLNRLIHRQILNSMATFLLKGQIRNGETVRVVVKDT 791
> SPBC4F6.17c
Length=803
Score = 93.2 bits (230), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 58/218 (26%), Positives = 107/218 (49%), Gaps = 47/218 (21%)
Query 1 FDEVENAHKNLWSLLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQFILEGYKEARAL 60
FDE+E AH ++ +LLL +LDEG L+DS+G +VDF + LI++TSN+G ++
Sbjct 610 FDELEKAHHDITNLLLQVLDEGFLTDSQGRKVDFRSTLIVMTSNLGSDILV--------- 660
Query 61 SGSNEKGGAADAAATGGAEDGSSGKGSGASSVGGFKGTGKSPKDVLRRMRQKVLKEVFGY 120
AD + T +PK R V+ V Y
Sbjct 661 ---------ADPSTTV------------------------TPKS-----RDAVMDVVQKY 682
Query 121 FKPQVIGRMSEIIIFEPLNDQALKGVLGLKLSSLTKGLAARGVDFKVADSAMTYILERAA 180
+ P+ + R+ + I+F L+++ L+ ++ ++L + + L R + V ++A ++ E+
Sbjct 683 YPPEFLNRIDDQIVFNKLSEKNLEDIVNVRLDEVQQRLNDRRIILTVTEAARKWLAEKGY 742
Query 181 SYKYGGRRMGKYLEKYIKPRVAPLLISGKLKRGDRAVL 218
S YG R + + ++K I +A +I G++K + V+
Sbjct 743 SPAYGARPLNRLIQKRILNTMAMKIIQGEIKSDENVVI 780
> At5g15450
Length=968
Score = 92.0 bits (227), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 54/215 (25%), Positives = 103/215 (47%), Gaps = 45/215 (20%)
Query 1 FDEVENAHKNLWSLLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQFILEGY-KEARA 59
FDE+E AH +++++ L +LD+G ++DS+G V FTN +II+TSN+G QFIL +A
Sbjct 756 FDEIEKAHGDVFNVFLQILDDGRVTDSQGRTVSFTNTVIIMTSNVGSQFILNNTDDDANE 815
Query 60 LSGSNEKGGAADAAATGGAEDGSSGKGSGASSVGGFKGTGKSPKDVLRRMRQKVLKEVFG 119
LS ++++V+
Sbjct 816 LSYET--------------------------------------------IKERVMNAARS 831
Query 120 YFKPQVIGRMSEIIIFEPLNDQALKGVLGLKLSSLTKGLAARGVDFKVADSAMTYILERA 179
F+P+ + R+ E I+F+PL+ + + ++ L+L+ + K +A R + + D+A+ +
Sbjct 832 IFRPEFMNRVDEYIVFKPLDREQINRIVRLQLARVQKRIADRKMKINITDAAVDLLGSLG 891
Query 180 ASYKYGGRRMGKYLEKYIKPRVAPLLISGKLKRGD 214
YG R + + +++ I+ +A ++ G K D
Sbjct 892 YDPNYGARPVKRVIQQNIENELAKGILRGDFKEED 926
> At3g48870
Length=952
Score = 90.9 bits (224), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 55/218 (25%), Positives = 103/218 (47%), Gaps = 36/218 (16%)
Query 1 FDEVENAHKNLWSLLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQFILEGYKEARAL 60
FDE+E AH ++++++L +L++G L+DSKG VDF N L+I+TSN+G I
Sbjct 737 FDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVI---------- 786
Query 61 SGSNEKGGAADAAATGGAEDGSSGKGSGASSVGGFKGTGKSPKDVLRRMRQKVLKEVFGY 120
EKGG E SS R++ V +E+ Y
Sbjct 787 ----EKGGRRIGFDLDHDEKDSS----------------------YNRIKSLVTEELKQY 820
Query 121 FKPQVIGRMSEIIIFEPLNDQALKGVLGLKLSSLTKGLAARGVDFKVADSAMTYILERAA 180
F+P+ + R+ E+I+F L +K + + L + L + ++ +V + +++
Sbjct 821 FRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVVARLEVKEIELQVTERFKERVVDEGF 880
Query 181 SYKYGGRRMGKYLEKYIKPRVAPLLISGKLKRGDRAVL 218
YG R + + + + ++ +A ++S +K GD ++
Sbjct 881 DPSYGARPLRRAIMRLLEDSMAEKMLSRDIKEGDSVIV 918
> At2g25140
Length=874
Score = 90.1 bits (222), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 50/218 (22%), Positives = 102/218 (46%), Gaps = 41/218 (18%)
Query 1 FDEVENAHKNLWSLLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQFILEGYKEARAL 60
FDE+E AH +++++LL +LD+G ++DS+G V F NC++I+TSNIG ILE +
Sbjct 671 FDEIEKAHPDVFNILLQLLDDGRITDSQGRTVSFKNCVVIMTSNIGSHHILETLR----- 725
Query 61 SGSNEKGGAADAAATGGAEDGSSGKGSGASSVGGFKGTGKSPKDVLRRMRQKVLKEVFGY 120
+NE A V M+++V++
Sbjct 726 --NNEDSKEA----------------------------------VYEIMKRQVVELARQN 749
Query 121 FKPQVIGRMSEIIIFEPLNDQALKGVLGLKLSSLTKGLAARGVDFKVADSAMTYILERAA 180
F+P+ + R+ E I+F+PL+ + ++ L++ + L + + + A+ + +
Sbjct 750 FRPEFMNRIDEYIVFQPLDSNEISKIVELQMRRVKNSLEQKKIKLQYTKEAVDLLAQLGF 809
Query 181 SYKYGGRRMGKYLEKYIKPRVAPLLISGKLKRGDRAVL 218
YG R + + +++ ++ +A ++ G D ++
Sbjct 810 DPNYGARPVKRVIQQMVENEIAVGILKGDFAEEDTVLV 847
> At5g50920
Length=929
Score = 89.4 bits (220), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 54/218 (24%), Positives = 106/218 (48%), Gaps = 36/218 (16%)
Query 1 FDEVENAHKNLWSLLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQFILEGYKEARAL 60
FDE+E AH ++++++L +L++G L+DSKG VDF N L+I+TSN+G I
Sbjct 716 FDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVI---------- 765
Query 61 SGSNEKGGAADAAATGGAEDGSSGKGSGASSVGGFKGTGKSPKDVLRRMRQKVLKEVFGY 120
EKGG E SS R++ V +E+ Y
Sbjct 766 ----EKGGRRIGFDLDYDEKDSS----------------------YNRIKSLVTEELKQY 799
Query 121 FKPQVIGRMSEIIIFEPLNDQALKGVLGLKLSSLTKGLAARGVDFKVADSAMTYILERAA 180
F+P+ + R+ E+I+F L +K + + L + + L + ++ +V + +++
Sbjct 800 FRPEFLNRLDEMIVFRQLTKLEVKEIADILLKEVFERLKKKEIELQVTERFKERVVDEGY 859
Query 181 SYKYGGRRMGKYLEKYIKPRVAPLLISGKLKRGDRAVL 218
+ YG R + + + + ++ +A +++ ++K GD ++
Sbjct 860 NPSYGARPLRRAIMRLLEDSMAEKMLAREIKEGDSVIV 897
> ECU11g1420
Length=851
Score = 82.0 bits (201), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 56/227 (24%), Positives = 101/227 (44%), Gaps = 52/227 (22%)
Query 1 FDEVENAHKNLWSLLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQFILEGYKEARAL 60
DEV+ AH+ + + L +LDEG ++D KG VDF NC++I+TSN+GQ+ I+
Sbjct 660 LDEVDLAHQTVLNTLYQLLDEGRVTDGKGAVVDFRNCVVIMTSNLGQEIIMR-------- 711
Query 61 SGSNEKGGAADAAATGGAEDGSSGKGSGASSVGGFKGTGKSPKDVLRRMRQKVLKEVFGY 120
+ G D R+K+ + V
Sbjct 712 -------------SRGELNDDE---------------------------RKKIEEIVLNR 731
Query 121 FKPQVIGRMSEIIIFEPLNDQALKGVLGLKLSSLTKGLAARGVDFKVADSAMTYILERAA 180
F + R+ E++ F L+ + + +L ++ L K L RG+ FKV+D+ ++ +
Sbjct 732 FGAPFVNRIDEVLYFNNLDRENMHRILEYQMGYLEKKLRGRGITFKVSDAVKEDMVTKVL 791
Query 181 SYKYGGRRMGKYLEKYIKPRVAPLLISGKLKRGDRAVLARSNTNPNQ 227
S YG R++ +++ K + +L L R + + S T+P++
Sbjct 792 SSIYGARQLRRFIRSSFKRALTKVL----LMRVNEDPIEVSCTHPSE 834
> SPBC16D10.08c
Length=905
Score = 75.1 bits (183), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 52/224 (23%), Positives = 104/224 (46%), Gaps = 46/224 (20%)
Query 1 FDEVENAHKNLWSLLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQFILEGYKEARAL 60
FDE+E A + ++LL +LD+G ++ +G VD N +II+TSN+G ++ L
Sbjct 692 FDEIEKAAPEVLTVLLQVLDDGRITSGQGQVVDAKNAVIIMTSNLGAEY----------L 741
Query 61 SGSNEKGGAADAAATGGAEDGSSGKGSGASSVGGFKGTGKSPKDVLRRMRQKVLKEVFGY 120
+ NE ++DG + R+ V+ + G+
Sbjct 742 TTDNE------------SDDGKIDSTT----------------------REMVMNSIRGF 767
Query 121 FKPQVIGRMSEIIIFEPLNDQALKGVLGLKLSSLTKGLAA--RGVDFKVADSAMTYILER 178
F+P+ + R+S I+IF L ++ ++ ++ + K L + R + +V+D A +
Sbjct 768 FRPEFLNRISSIVIFNRLRRVDIRNIVENRILEVQKRLQSNHRSIKIEVSDEAKDLLGSA 827
Query 179 AASYKYGGRRMGKYLEKYIKPRVAPLLISGKLKRGDRAVLARSN 222
S YG R + + ++ + +A L+++G+L+ + A + N
Sbjct 828 GYSPAYGARPLNRVIQNQVLNPMAVLILNGQLRDKETAHVVVQN 871
> YLL026w
Length=908
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 48/213 (22%), Positives = 99/213 (46%), Gaps = 49/213 (23%)
Query 1 FDEVENAHKNLWSLLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQFILEGYKEARAL 60
FDEVE AH ++ +++L MLD+G ++ +G +D +NC++I+TSN+G +FI
Sbjct 685 FDEVEKAHPDVLTVMLQMLDDGRITSGQGKTIDCSNCIVIMTSNLGAEFI---------- 734
Query 61 SGSNEKGGAADAAATGGAEDGSSGKGSGASSVGGFKGTGKSPKDVLRRMRQKVLKEVFGY 120
++ GS + S + V G +RQ +
Sbjct 735 ----------------NSQQGSKIQESTKNLVMG-------------AVRQ--------H 757
Query 121 FKPQVIGRMSEIIIFEPLNDQALKGVLGLKLSSLTKGLAARGVDFK--VADSAMTYILER 178
F+P+ + R+S I+IF L+ +A+ ++ ++L + + +K + A ++ +
Sbjct 758 FRPEFLNRISSIVIFNKLSRKAIHKIVDIRLKEIEERFEQNDKHYKLNLTQEAKDFLAKY 817
Query 179 AASYKYGGRRMGKYLEKYIKPRVAPLLISGKLK 211
S G R + + ++ I ++A ++ ++K
Sbjct 818 GYSDDMGARPLNRLIQNEILNKLALRILKNEIK 850
> Hs13540606
Length=707
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 45/208 (21%), Positives = 94/208 (45%), Gaps = 31/208 (14%)
Query 1 FDEVENAHKNLWSLLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQ----QFILEGYKE 56
FDEV+ AH ++ +++L + DEG L+D KG +D + + I+TSN+ Q L+ +E
Sbjct 453 FDEVDKAHPDVLTIMLQLFDEGRLTDGKGKTIDCKDAIFIMTSNVASDEIAQHALQLRQE 512
Query 57 ARALSGSNEKGGAADAAATGGAEDGSSGKGSGASSVGGFKGTGKSPKDVLRRMRQKVLKE 116
A +S + A ++G + + K + + ++ V++
Sbjct 513 ALEMS-----------------------RNRIAENLGDVQISDKI--TISKNFKENVIRP 547
Query 117 VF--GYFKPQVIGRMSEIIIFEPLNDQALKGVLGLKLSSLTKGLAARGVDFKVADSAMTY 174
+ + + + +GR++EI+ F P L ++ +L+ K R + D +
Sbjct 548 ILKAHFRRDEFLGRINEIVYFLPFCHSELIQLVNKELNFWAKRAKQRHNITLLWDREVAD 607
Query 175 ILERAASYKYGGRRMGKYLEKYIKPRVA 202
+L + YG R + +E+ + ++A
Sbjct 608 VLVDGYNVHYGARSIKHEVERRVVNQLA 635
> At2g25030
Length=265
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 44/216 (20%), Positives = 85/216 (39%), Gaps = 63/216 (29%)
Query 1 FDEVENAHKNLWSLLLPMLDEGHLSDSKGNRVDFTNCLIILTSNIGQQFILEGYKEARAL 60
FDE+E H +++S+LL +LD+G +T++ G ILE +
Sbjct 83 FDEIEKPHLDVFSILLQLLDDGR-----------------ITNSHGSLHILETIR----- 120
Query 61 SGSNEKGGAADAAATGGAEDGSSGKGSGASSVGGFKGTGKSPKDVLRRMRQKVLKEVFGY 120
+NE A M+Q+V++
Sbjct 121 --NNEDIKEA----------------------------------FYEMMKQQVVELARKT 144
Query 121 FKPQVIGRMSEIIIFEPLNDQALKGVLGLKLSSLTKGLAARGVDFKVADSAMTYILERAA 180
F+P+ + R+ E I+ +PLN + ++ L++ + K L ++ + A+ + +
Sbjct 145 FRPKFMNRIDEYIVSQPLNSSEISKIVELQMRQVKKRLEQNKINLEYTKEAVDLLAQLGF 204
Query 181 SYKYGGRRMGKYLEKYIKPRVAPLLISGKLKRGDRA 216
G R + + +EK +K I+ K+ +GD A
Sbjct 205 DPNNGARPVKQMIEKLVKKE-----ITLKVLKGDFA 235
> At5g13680
Length=1319
Score = 35.0 bits (79), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 38/143 (26%), Positives = 65/143 (45%), Gaps = 17/143 (11%)
Query 84 GKGSGASSVGGFKGTGKSPKDVLRRMRQKVLKEVFGYFKPQVIGRMSEIIIFEPLNDQAL 143
G SG VG GK D + ++ ++ +EV KP + I E +D +
Sbjct 1025 GDWSGVLRVGALMKLGK---DEILKLAYELCEEVNALGKPAEAAK----IALEYCSD--I 1075
Query 144 KGVLGLKLSSLTKGLAARGVDFKVADSAMTYI----LERAA----SYKYGGRRMGKYLEK 195
G + L +++ A R AD ++ + LE A+ +K ++GKYL +
Sbjct 1076 SGGISLLINAREWEEALRVAFLHTADDRISVVKSSALECASGLVSEFKESIEKVGKYLTR 1135
Query 196 YIKPRVAPLLISGKLKRGDRAVL 218
Y+ R LL++ KLK +R+V+
Sbjct 1136 YLAVRQRRLLLAAKLKSEERSVV 1158
> Hs18595579
Length=239
Score = 33.5 bits (75), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 36/80 (45%), Gaps = 4/80 (5%)
Query 89 ASSVGGFKGTGKSPKDVLRRMRQKVLKEVFGYFKPQVIGRMSEIIIFEPLN--DQALKGV 146
A+S GG G +P R Q+ L VFG Q I S+ ++ L + +KG+
Sbjct 74 AASTGGHPGCCGTPVSSCRPKEQEALNSVFGQHDSQPIFPTSQYVLGHILGLVEMRMKGM 133
Query 147 LGLKLSSLTKGLAARGVDFK 166
L L SL ARG D K
Sbjct 134 --LSLGSLRHCKTARGQDQK 151
> 7302267
Length=1486
Score = 30.0 bits (66), Expect = 5.3, Method: Composition-based stats.
Identities = 19/71 (26%), Positives = 34/71 (47%), Gaps = 4/71 (5%)
Query 59 ALSGSNEKGGAADAAATGGAEDGSSGKGSGASSVGGFKGTGKSPKDVLRRMRQKVLKE-- 116
A + N++ A AA G + +GS G GA+S + D +R++ ++E
Sbjct 84 ASANKNKRTAAGSGAALGLSGEGSCLTGGGAASAKKARSDLPGSFDASKRLKVAAMEESQ 143
Query 117 --VFGYFKPQV 125
+ G+FK Q+
Sbjct 144 TKITGFFKSQM 154
Lambda K H
0.314 0.135 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5737723452
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40