bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0851_orf1
Length=97
Score E
Sequences producing significant alignments: (Bits) Value
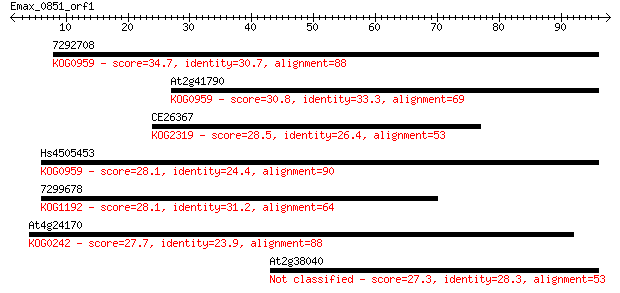
7292708 34.7 0.051
At2g41790 30.8 0.70
CE26367 28.5 3.1
Hs4505453 28.1 4.2
7299678 28.1 4.2
At4g24170 27.7 6.1
At2g38040 27.3 6.5
> 7292708
Length=1077
Score = 34.7 bits (78), Expect = 0.051, Method: Composition-based stats.
Identities = 27/104 (25%), Positives = 49/104 (47%), Gaps = 17/104 (16%)
Query 8 IFWSAPANLETQLVEL-----------QCQPSSLLQYLLEHPVKGGLLETLKEKEYISK- 55
+F+ P ET+L EL + +P L YLL + +G L L+ + + +
Sbjct 311 VFFVKPVENETKL-ELTWVLPNVRQYYRSKPDQFLSYLLGYEGRGSLCAYLRRRLWALQL 369
Query 56 ----GEAFTDMTSRSSILGLVLDLTEKGESNVTELLGSVHAHIQ 95
E DM S S+ + + LT++G N+ E+L + A+++
Sbjct 370 IAGIDENGFDMNSMYSLFNICIYLTDEGFKNLDEVLAATFAYVK 413
> At2g41790
Length=970
Score = 30.8 bits (68), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 37/72 (51%), Gaps = 5/72 (6%)
Query 27 PSSLLQYLLEHPVKGGLLETLKEKEY---ISKGEAFTDMTSRSSILGLVLDLTEKGESNV 83
PS L +L+ H +G L LK + +S GE + T S + +DLT+ G ++
Sbjct 291 PSQYLGHLIGHEGEGSLFHALKTLGWATGLSAGEG--EWTLDYSFFKVSIDLTDAGHEHM 348
Query 84 TELLGSVHAHIQ 95
E+LG + +IQ
Sbjct 349 QEILGLLFNYIQ 360
> CE26367
Length=1109
Score = 28.5 bits (62), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 14/53 (26%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 24 QCQPSSLLQYLLEHPVKGGLLETLKEKEYISKGEAFTDMTSRSSILGLVLDLT 76
QC+ + L+ +++ + L ++ I+ G A +DM ++SS +G V D+
Sbjct 371 QCRNTHLISEMMDALLVEKLAPDVEITSLIANGTAESDMATKSSFVGTVADVN 423
> Hs4505453
Length=1219
Score = 28.1 bits (61), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 22/95 (23%), Positives = 44/95 (46%), Gaps = 9/95 (9%)
Query 6 LFIFWSAPANLETQLVELQCQPSSLLQYLLEHPVKGGLLETLKEKEYI-----SKGEAFT 60
L I W+ P Q + +P + +L+ H KG +L L++K + GE
Sbjct 508 LTITWALPP----QQQHYRVKPLHYISWLVGHEGKGSILSFLRKKCWALALFGGNGETGF 563
Query 61 DMTSRSSILGLVLDLTEKGESNVTELLGSVHAHIQ 95
+ S S+ + + LT++G + E+ +V +++
Sbjct 564 EQNSTYSVFSISITLTDEGYEHFYEVAYTVFLYLK 598
> 7299678
Length=530
Score = 28.1 bits (61), Expect = 4.2, Method: Composition-based stats.
Identities = 20/70 (28%), Positives = 32/70 (45%), Gaps = 7/70 (10%)
Query 6 LFIFWSAPANLETQLVELQCQPSSLLQYLLEHPVKGGLLETLKEKEYISKGEAFTD---- 61
L +F P N + + + SLL L E P K +LE L +Y K +F+
Sbjct 394 LPVFGDQPGNADVMVKQGFGLTQSLLS-LEEQPFKEAILEILSNPQYFDKVASFSSLYRD 452
Query 62 --MTSRSSIL 69
M++R S++
Sbjct 453 RPMSARESVI 462
> At4g24170
Length=1263
Score = 27.7 bits (60), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 21/88 (23%), Positives = 40/88 (45%), Gaps = 7/88 (7%)
Query 4 RRLFIFWSAPANLETQLVELQCQPSSLLQYLLEHPVKGGLLETLKEKEYISKGEAFTDMT 63
R+ + +SA + +L C+ SS LL+ P +G ++E L+E E D +
Sbjct 121 RKFTLKFSAMEIYNEAVRDLLCEDSSTPLRLLDDPERGTVVEKLRE-------ETLRDRS 173
Query 64 SRSSILGLVLDLTEKGESNVTELLGSVH 91
+L + + GE+++ E+ H
Sbjct 174 HLEELLSICETQRKIGETSLNEISSRSH 201
> At2g38040
Length=796
Score = 27.3 bits (59), Expect = 6.5, Method: Composition-based stats.
Identities = 15/53 (28%), Positives = 28/53 (52%), Gaps = 0/53 (0%)
Query 43 LLETLKEKEYISKGEAFTDMTSRSSILGLVLDLTEKGESNVTELLGSVHAHIQ 95
L + LKEK +KGE +M +GL LD ++ + + E + + + ++Q
Sbjct 624 LPDALKEKVLKTKGEVEAEMAGVLKSMGLELDAVKQNQKDTAEQIYAANENLQ 676
Lambda K H
0.316 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1198045380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40