bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
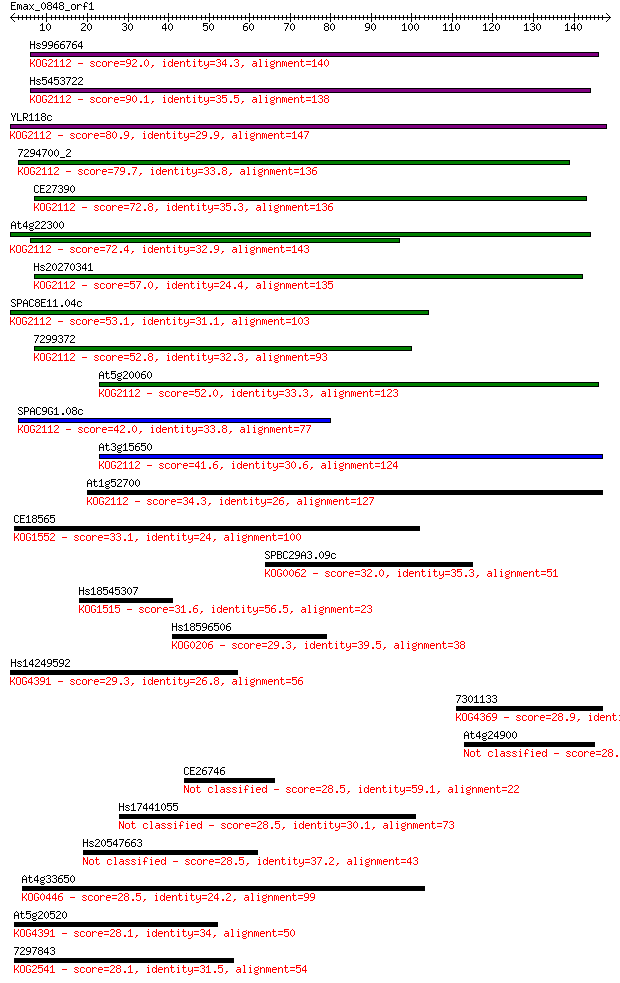
Query= Emax_0848_orf1
Length=148
Score E
Sequences producing significant alignments: (Bits) Value
Hs9966764 92.0 4e-19
Hs5453722 90.1 1e-18
YLR118c 80.9 7e-16
7294700_2 79.7 2e-15
CE27390 72.8 2e-13
At4g22300 72.4 3e-13
Hs20270341 57.0 1e-08
SPAC8E11.04c 53.1 2e-07
7299372 52.8 2e-07
At5g20060 52.0 5e-07
SPAC9G1.08c 42.0 4e-04
At3g15650 41.6 6e-04
At1g52700 34.3 0.089
CE18565 33.1 0.18
SPBC29A3.09c 32.0 0.44
Hs18545307 31.6 0.55
Hs18596506 29.3 2.7
Hs14249592 29.3 3.2
7301133 28.9 3.4
At4g24900 28.9 3.8
CE26746 28.5 4.5
Hs17441055 28.5 5.0
Hs20547663 28.5 5.4
At4g33650 28.5 5.5
At5g20520 28.1 5.7
7297843 28.1 6.3
> Hs9966764
Length=231
Score = 92.0 bits (227), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 48/140 (34%), Positives = 76/140 (54%), Gaps = 9/140 (6%)
Query 6 RIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVIEV 65
I +I+ +++ GI +RI++GGFSQGGA++ LT PH L GI+A+S W PL +
Sbjct 98 NIKALIEHEMKNGIPANRIVLGGFSQGGALSLYTALTCPHPLAGIVALSCWLPLHRAFPQ 157
Query 66 SPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYPGL 125
+ + I+ CHG +D MV +G + E +R+ + A + FK YPG+
Sbjct 158 AANGSAKDLAILQCHGELDPMVPVRFGALTAEKLRSVVTPA---------RVQFKTYPGV 208
Query 126 GHFANQQELNDIKAFLSRTF 145
H + QE+ +K FL +
Sbjct 209 MHSSCPQEMAAVKEFLEKLL 228
> Hs5453722
Length=230
Score = 90.1 bits (222), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 49/141 (34%), Positives = 78/141 (55%), Gaps = 14/141 (9%)
Query 6 RIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVIEV 65
I +ID +++ GI +RI++GGFSQGGA++ LT+ KL G+ A+S W PL
Sbjct 95 NIKALIDQEVKNGIPSNRIILGGFSQGGALSLYTALTTQQKLAGVTALSCWLPLRASFPQ 154
Query 66 SPHYVEAVPR---IMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLY 122
P + R I+ CHG D +V ++G +VE ++ + A ++TFK Y
Sbjct 155 GP--IGGANRDISILQCHGDCDPLVPLMFGSLTVEKLKTLVNPA---------NVTFKTY 203
Query 123 PGLGHFANQQELNDIKAFLSR 143
G+ H + QQE+ D+K F+ +
Sbjct 204 EGMMHSSCQQEMMDVKQFIDK 224
> YLR118c
Length=227
Score = 80.9 bits (198), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 44/148 (29%), Positives = 80/148 (54%), Gaps = 11/148 (7%)
Query 1 MDSKARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLG 60
M+S I+K + +I+ GI P +I++GGFSQG A+A +T P K+GGI+A+S +C +
Sbjct 90 MNSLNSIEKTVKQEIDKGIKPEQIIIGGFSQGAALALATSVTLPWKIGGIVALSGFCSIP 149
Query 61 KVIEVSPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQ-EDITF 119
+++ + + I H HG +D +V G + + + + C+ ++ F
Sbjct 150 GILKQHKNGINVKTPIFHGHGDMDPVVPIGLGIKAKQFYQ----------DSCEIQNYEF 199
Query 120 KLYPGLGHFANQQELNDIKAFLSRTFGS 147
K+Y G+ H EL D+ +F+ ++ S
Sbjct 200 KVYKGMAHSTVPDELEDLASFIKKSLSS 227
> 7294700_2
Length=211
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 46/136 (33%), Positives = 74/136 (54%), Gaps = 12/136 (8%)
Query 3 SKARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKV 62
++ + +I +I AGI +RI++GGFSQGGA+A LT L G++A+S W PL K
Sbjct 59 ARDSVHGMIQKEISAGIPANRIVLGGFSQGGALALYSALTYDQPLAGVVALSCWLPLHKQ 118
Query 63 IEVSPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLY 122
+ + VP I HG D +V +GQ S +++ + +++TFK Y
Sbjct 119 FPGAKVNSDDVP-IFQAHGDYDPVVPYKFGQLSASLLKSFM-----------KNVTFKTY 166
Query 123 PGLGHFANQQELNDIK 138
GL H ++ E++D+K
Sbjct 167 SGLSHSSSDDEMDDVK 182
> CE27390
Length=223
Score = 72.8 bits (177), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 48/136 (35%), Positives = 69/136 (50%), Gaps = 12/136 (8%)
Query 7 IDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVIEVS 66
+ ++ID+++ AGI SRI VGGFS GGA+A LT P KLGGI+ +SS+ S
Sbjct 95 VHQLIDAEVAAGIPASRIAVGGFSMGGALAIYAGLTYPQKLGGIVGLSSFFLQRTKFPGS 154
Query 67 PHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYPGLG 126
A P I HG D +V +GQ S + ++ K + Y G+
Sbjct 155 FTANNATP-IFLGHGTDDFLVPLQFGQMSEQYIK-----------KFNPKVELHTYRGMQ 202
Query 127 HFANQQELNDIKAFLS 142
H + +E+ D+K FLS
Sbjct 203 HSSCGEEMRDVKTFLS 218
> At4g22300
Length=471
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 47/152 (30%), Positives = 71/152 (46%), Gaps = 20/152 (13%)
Query 1 MDSKARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPL- 59
+++ + IID +I G +P + + G SQGGA+ L P LGG +S W P
Sbjct 77 LEAVKNVHAIIDQEIAEGTNPENVFICGLSQGGALTLASVLLYPKTLGGGAVLSGWVPFT 136
Query 60 GKVIEVSPHYVEAVPR--------IMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPE 111
+I P + VP I+ HG D MV GQ ++ ++ +AG E
Sbjct 137 SSIISQFPEEAKKVPHLCFLINTPILWSHGTDDRMVLFEAGQAALPFLK----EAGVTCE 192
Query 112 KCQEDITFKLYPGLGHFANQQELNDIKAFLSR 143
FK YPGLGH + +EL I++++ R
Sbjct 193 -------FKAYPGLGHSISNKELKYIESWIKR 217
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 43/93 (46%), Gaps = 4/93 (4%)
Query 6 RIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVIEV 65
+ IID +I GI+P + + GFSQGGA+ L P +GG S W P I
Sbjct 367 NVHAIIDKEIAGGINPENVYICGFSQGGALTLASVLLYPKTIGGGAVFSGWIPFNSSI-- 424
Query 66 SPHYVEAVPR--IMHCHGAVDDMVRPVYGQTSV 96
+ + E + I+ HG D V GQ ++
Sbjct 425 TNQFTEDAKKTPILWSHGIDDKTVLFEAGQAAL 457
> Hs20270341
Length=237
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 33/136 (24%), Positives = 67/136 (49%), Gaps = 12/136 (8%)
Query 7 IDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVIEVS 66
+ +ID ++++GI +RIL+GGFS GG +A + + + G+ A+SS+ + +
Sbjct 101 LTDLIDEEVKSGIKKNRILIGGFSMGGCMAMHLAYRNHQDVAGVFALSSFLNKASAVYQA 160
Query 67 PHYVEAV-PRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYPGL 125
V P + CHG D++V + + + N ++ + K F +P +
Sbjct 161 LQKSNGVLPELFQCHGTADELVLHSWAEET-----NSMLKSLGVTTK------FHSFPNV 209
Query 126 GHFANQQELNDIKAFL 141
H ++ EL+ +K ++
Sbjct 210 YHELSKTELDILKLWI 225
> SPAC8E11.04c
Length=224
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 32/103 (31%), Positives = 51/103 (49%), Gaps = 0/103 (0%)
Query 1 MDSKARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLG 60
+ S ++ ++ID+++ GI RIL+GGFSQG V+ LT P +L GI+ S + PL
Sbjct 87 LRSAGQLHELIDAELALGIPSDRILIGGFSQGCMVSLYAGLTYPKRLAGIMGHSGFLPLA 146
Query 61 KVIEVSPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRL 103
+ V I+ + D +V V S + + N L
Sbjct 147 SKFPSALSRVAKEIPILLTYMTEDPIVPSVLSSASAKYLINNL 189
> 7299372
Length=235
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 52/95 (54%), Gaps = 2/95 (2%)
Query 7 IDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVI--E 64
++++ID ++ +GI +RI+VGGFS GGA+A L G+ A SS+ G V+
Sbjct 95 VNQLIDEEVASGIPLNRIVVGGFSMGGALALHTGYHLRRSLAGVFAHSSFLNRGSVVYDS 154
Query 65 VSPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESV 99
++ E+ P + HG D +V +G + E++
Sbjct 155 LANGKDESFPELRMYHGERDTLVPKDWGLETFENL 189
> At5g20060
Length=252
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 41/140 (29%), Positives = 64/140 (45%), Gaps = 28/140 (20%)
Query 23 RILVGGFSQGGAVAYL--VCLT---------SPHKLGGILAMSSWCPLGKVI------EV 65
++ VGGFS G A + C P L I+ +S W P K + E
Sbjct 120 KLGVGGFSMGAATSLYSATCFALGKYGNGNPYPINLSAIIGLSGWLPCAKTLAGKLEEEQ 179
Query 66 SPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYPGL 125
+ ++P I+ CHG DD+V +G+ S ++ L+ G + +TFK Y L
Sbjct 180 IKNRAASLP-IVVCHGKADDVVPFKFGEKSSQA----LLSNGFK------KVTFKPYSAL 228
Query 126 GHFANQQELNDIKAFLSRTF 145
GH QEL+++ A+L+ T
Sbjct 229 GHHTIPQELDELCAWLTSTL 248
> SPAC9G1.08c
Length=241
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 38/81 (46%), Gaps = 4/81 (4%)
Query 3 SKARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVC--LTSPHKLGGILAMSSWCPLG 60
S I +I + + GI SRI GF QG VA C L++ ++LGGI + PL
Sbjct 97 SFTMISNLIGNLLSYGILSSRIFFFGFGQGAMVALYSCYKLSTKYQLGGIFSFGGTLPLS 156
Query 61 KVIEVSPHYVEA--VPRIMHC 79
+ P +V + +HC
Sbjct 157 ITLPNHPFHVPVYLFEKRLHC 177
> At3g15650
Length=255
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 38/143 (26%), Positives = 60/143 (41%), Gaps = 31/143 (21%)
Query 23 RILVGGFSQGGAVAYLVCLTSPHKLG-------------GILAMSSWCPLGKVIEVSPHY 69
++ +GGFS G A+A + T+ + LG + +S W P + +
Sbjct 120 KVGIGGFSMGAAIA--LYSTTCYALGRYGTGHAYTINLRATVGLSGWLPGWRSLRSKIES 177
Query 70 VEAVPR------IMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYP 123
V R I+ HG DD+V +G+ S S L AG R FK Y
Sbjct 178 SNEVARRAASIPILLAHGTSDDVVPYRFGEKSAHS----LAMAGFR------QTMFKPYE 227
Query 124 GLGHFANQQELNDIKAFLSRTFG 146
GLGH+ +E++++ +L G
Sbjct 228 GLGHYTVPKEMDEVVHWLVSRLG 250
> At1g52700
Length=223
Score = 34.3 bits (77), Expect = 0.089, Method: Compositional matrix adjust.
Identities = 33/138 (23%), Positives = 54/138 (39%), Gaps = 45/138 (32%)
Query 20 DPSRILVGGFSQGGAVA-----------YLVCLTSPHKLGGILAMSSWCPLGKVIEVSPH 68
+P+ + +GGFS G A++ Y P L ++ +S W P
Sbjct 115 EPADVGIGGFSMGAAISLYSATCYALGRYGTGHAYPINLQAVVGLSGWLP---------- 164
Query 69 YVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNRLIDAGARPEKCQEDITFKLYPGLGHF 128
DD+V +G+ S +S L AG R FK Y GLGH+
Sbjct 165 --------------ADDVVPYRFGEKSAQS----LGMAGFRLA------MFKPYEGLGHY 200
Query 129 ANQQELNDIKAFLSRTFG 146
+E++++ +L+ G
Sbjct 201 TVPREMDEVVHWLTTMLG 218
> CE18565
Length=305
Score = 33.1 bits (74), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 24/115 (20%), Positives = 52/115 (45%), Gaps = 20/115 (17%)
Query 2 DSKARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGK 61
D +A +KI++ + + +I+V G+S G A + T+P +L G++ ++ + +
Sbjct 135 DVRAVYEKILEMRPD-----KKIVVMGYSIGTTAAVDLAATNPDRLAGVVLIAPFTSGLR 189
Query 62 VIEVSPHYVEAV---------------PRIMHCHGAVDDMVRPVYGQTSVESVRN 101
+ P + R++ CHG VD+++ +G E ++N
Sbjct 190 LFSSKPDKPDTCWADSFKSFDKINNIDTRVLICHGDVDEVIPLSHGLALYEKLKN 244
> SPBC29A3.09c
Length=736
Score = 32.0 bits (71), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 27/53 (50%), Gaps = 8/53 (15%)
Query 64 EVSPHYVEAVPRIMHCHGAVDDMVRPV--YGQTSVESVRNRLIDAGARPEKCQ 114
EV PH + +C G +D ++ V ++ +RN L+DAGAR E Q
Sbjct 11 EVDPHVLS------YCTGYANDAIKDVDEASDHTISLIRNLLLDAGAREEAVQ 57
> Hs18545307
Length=350
Score = 31.6 bits (70), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 13/23 (56%), Positives = 19/23 (82%), Gaps = 0/23 (0%)
Query 18 GIDPSRILVGGFSQGGAVAYLVC 40
G+DP+R++V G S GGA+A +VC
Sbjct 124 GVDPARVVVCGDSFGGAIAAVVC 146
> Hs18596506
Length=950
Score = 29.3 bits (64), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 20/38 (52%), Gaps = 6/38 (15%)
Query 41 LTSPHKLGGILAMSSWCPLGKVIEVSPHYVEAVPRIMH 78
L PH+LG +L SSW PL + + + P PR H
Sbjct 898 LICPHQLGLLLPSSSWLPLPEDLGLGP------PRYYH 929
> Hs14249592
Length=201
Score = 29.3 bits (64), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 15/56 (26%), Positives = 32/56 (57%), Gaps = 3/56 (5%)
Query 1 MDSKARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSW 56
+DS+A +D ++ +D ++I + G S GGAVA + + H++ I+ +++
Sbjct 31 LDSEAVLDYVM---TRPDLDKTKIFLFGRSLGGAVAIHLASENSHRISAIMVENTF 83
> 7301133
Length=2441
Score = 28.9 bits (63), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 22/39 (56%), Gaps = 3/39 (7%)
Query 111 EKCQED---ITFKLYPGLGHFANQQELNDIKAFLSRTFG 146
++C++D +LY GL HFAN E+ DI L+ +
Sbjct 304 QQCEDDGSASASELYSGLEHFANDGEMEDIFQELASSLN 342
> At4g24900
Length=423
Score = 28.9 bits (63), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 19/37 (51%), Gaps = 5/37 (13%)
Query 113 CQEDI-----TFKLYPGLGHFANQQELNDIKAFLSRT 144
C EDI +F + HFA+ L +IK FLS+
Sbjct 75 CDEDIVELGSSFACSKAINHFASSDHLKNIKQFLSKN 111
> CE26746
Length=367
Score = 28.5 bits (62), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 13/22 (59%), Positives = 14/22 (63%), Gaps = 0/22 (0%)
Query 44 PHKLGGILAMSSWCPLGKVIEV 65
P K G +L SSW PL K IEV
Sbjct 113 PSKSGPVLLSSSWLPLNKDIEV 134
> Hs17441055
Length=112
Score = 28.5 bits (62), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 22/80 (27%), Positives = 37/80 (46%), Gaps = 11/80 (13%)
Query 28 GFSQGGAVAYLVCLTSP-----HKLGGILAMSSWCPLGKVIEVS--PHYVEAVPRIMHCH 80
GF QGG A + +T P H + LA ++ P+ + + S P +++ P HC
Sbjct 3 GFKQGGPRAAIALITHPLSPSLHPIAASLAPAAIVPMPRPLCPSSVPSRMDSCP---HC- 58
Query 81 GAVDDMVRPVYGQTSVESVR 100
++ + P YG T + R
Sbjct 59 PSLKPSLAPKYGSTKIAGKR 78
> Hs20547663
Length=282
Score = 28.5 bits (62), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 23/48 (47%), Gaps = 5/48 (10%)
Query 19 IDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSS-----WCPLGK 61
+DP R+ + G S GG A + L +P K + A + CP GK
Sbjct 138 VDPQRMSIFGHSMGGHGALICALKNPGKYKSVSAFAPICNPVLCPWGK 185
> At4g33650
Length=808
Score = 28.5 bits (62), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 24/99 (24%), Positives = 45/99 (45%), Gaps = 13/99 (13%)
Query 4 KARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGILAMSSWCPLGKVI 63
+ R+D++I + G++PS ++G + Y + + P+ +GG K +
Sbjct 489 RKRMDEVIGDFLREGLEPSEAMIGDIID-MEMDY-INTSHPNFIGGT----------KAV 536
Query 64 EVSPHYVEAVPRIMHCHGAVDDMVRPVYGQTSVESVRNR 102
E + H V++ RI H D V P +S V++R
Sbjct 537 EAAMHQVKS-SRIPHPVARPKDTVEPDRTSSSTSQVKSR 574
> At5g20520
Length=340
Score = 28.1 bits (61), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 28/50 (56%), Gaps = 3/50 (6%)
Query 2 DSKARIDKIIDSQIEAGIDPSRILVGGFSQGGAVAYLVCLTSPHKLGGIL 51
D++A +D + ID SRI+V G S GGAV ++ +P K+ ++
Sbjct 161 DAQAALDHLSG---RTDIDTSRIVVFGRSLGGAVGAVLTKNNPDKVSALI 207
> 7297843
Length=288
Score = 28.1 bits (61), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 32/56 (57%), Gaps = 2/56 (3%)
Query 2 DSKARIDKIIDSQIEAG-IDPSRILVGGFSQGGAVAYLVCLTSP-HKLGGILAMSS 55
++ ++D++ D E G + P I+V G+SQGG +A + P H + +++SS
Sbjct 69 NAWRQVDQVRDYLNEVGKLHPEGIIVLGYSQGGLLARAAIQSLPEHNVKTFISLSS 124
Lambda K H
0.321 0.138 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1821716300
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40