bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
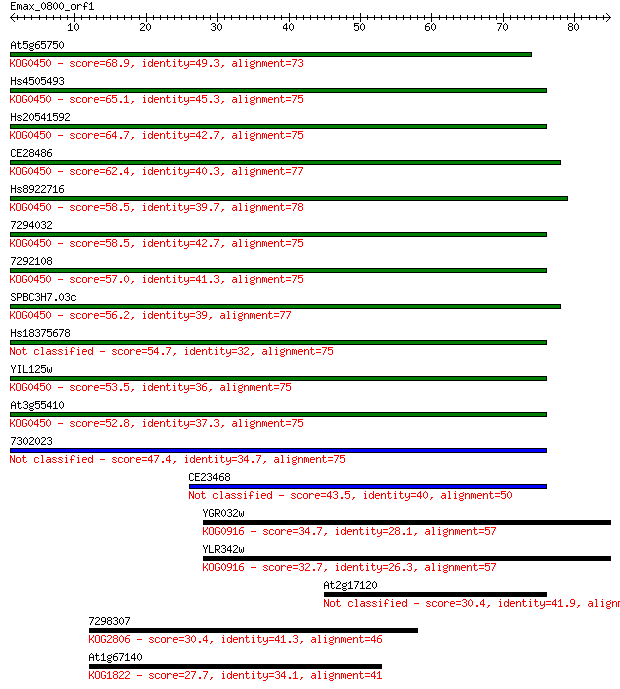
Query= Emax_0800_orf1
Length=84
Score E
Sequences producing significant alignments: (Bits) Value
At5g65750 68.9 2e-12
Hs4505493 65.1 3e-11
Hs20541592 64.7 4e-11
CE28486 62.4 2e-10
Hs8922716 58.5 3e-09
7294032 58.5 3e-09
7292108 57.0 7e-09
SPBC3H7.03c 56.2 1e-08
Hs18375678 54.7 4e-08
YIL125w 53.5 1e-07
At3g55410 52.8 2e-07
7302023 47.4 7e-06
CE23468 43.5 1e-04
YGR032w 34.7 0.040
YLR342w 32.7 0.18
At2g17120 30.4 0.76
7298307 30.4 0.84
At1g67140 27.7 4.9
> At5g65750
Length=1025
Score = 68.9 bits (167), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 36/73 (49%), Positives = 48/73 (65%), Gaps = 10/73 (13%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G++YY+L + ER +SE D VA+ RVEQL PFP+DL R+++RYPN +VW
Sbjct 903 GKVYYEL--DEERKKSETKD-------VAICRVEQLCPFPYDLIQRELKRYPNAE-IVWC 952
Query 61 QEEPMNQGKERRI 73
QEEPMN G + I
Sbjct 953 QEEPMNMGGYQYI 965
> Hs4505493
Length=1002
Score = 65.1 bits (157), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 34/75 (45%), Positives = 47/75 (62%), Gaps = 9/75 (12%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G++YYDL ER+ D G +VA+ R+EQLSPFPFDL ++++Q+YPN + W
Sbjct 906 GKVYYDLTRERKA-------RDMVG-QVAITRIEQLSPFPFDLLLKEVQKYPNAE-LAWC 956
Query 61 QEEPMNQGKERRINP 75
QEE NQG + P
Sbjct 957 QEEHKNQGYYDYVKP 971
> Hs20541592
Length=1023
Score = 64.7 bits (156), Expect = 4e-11, Method: Composition-based stats.
Identities = 32/75 (42%), Positives = 46/75 (61%), Gaps = 9/75 (12%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G++YYDL ER+ + +VA+ R+EQLSPFPFDL ++++Q+YPN + W
Sbjct 906 GKVYYDLTRERK--------ARDMVGQVAITRIEQLSPFPFDLLLKEVQKYPN-AELAWC 956
Query 61 QEEPMNQGKERRINP 75
QEE NQG + P
Sbjct 957 QEEHKNQGYYDYVKP 971
> CE28486
Length=1029
Score = 62.4 bits (150), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 31/77 (40%), Positives = 47/77 (61%), Gaps = 9/77 (11%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G++YYD+++ R+ + EN+ VAL RVEQLSPFP+DL ++ ++Y ++WA
Sbjct 908 GKVYYDMVAARKHVGKEND--------VALVRVEQLSPFPYDLVQQECRKYQGAE-ILWA 958
Query 61 QEEPMNQGKERRINPII 77
QEE N G + P I
Sbjct 959 QEEHKNMGAWSFVQPRI 975
> Hs8922716
Length=1010
Score = 58.5 bits (140), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 31/78 (39%), Positives = 44/78 (56%), Gaps = 9/78 (11%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G++YYDL+ ER E +VA+ R+EQ+SPFPFDL ++ ++YP + W
Sbjct 892 GKVYYDLVKERSSQDLEE--------KVAITRLEQISPFPFDLIKQEAEKYPGAE-LAWC 942
Query 61 QEEPMNQGKERRINPIIM 78
QEE N G I+P M
Sbjct 943 QEEHKNMGYYDYISPRFM 960
> 7294032
Length=1001
Score = 58.5 bits (140), Expect = 3e-09, Method: Composition-based stats.
Identities = 32/75 (42%), Positives = 39/75 (52%), Gaps = 9/75 (12%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G++YYDL R Q E +A+ RVEQ+SPFPFDL Y N +VWA
Sbjct 898 GRVYYDLTKTRREKQLEG--------EIAIVRVEQISPFPFDLVKEQANLYKN-AELVWA 948
Query 61 QEEPMNQGKERRINP 75
QEE NQG + P
Sbjct 949 QEEHKNQGSWTYVQP 963
> 7292108
Length=1075
Score = 57.0 bits (136), Expect = 7e-09, Method: Composition-based stats.
Identities = 31/75 (41%), Positives = 41/75 (54%), Gaps = 9/75 (12%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G++YYDL+ ER D VAL RVEQL PFP+DL + ++ YP ++WA
Sbjct 922 GKVYYDLVKER--------DDHEQVETVALVRVEQLCPFPYDLISQQLELYPK-AELLWA 972
Query 61 QEEPMNQGKERRINP 75
QEE N G + P
Sbjct 973 QEEHKNMGAWSYVQP 987
> SPBC3H7.03c
Length=1009
Score = 56.2 bits (134), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 30/77 (38%), Positives = 42/77 (54%), Gaps = 9/77 (11%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
GQ++ L RE N DN +A+ RVEQL PF + +I +YPNL+ ++W
Sbjct 897 GQVWVALSKAREE-----NKIDN----IAITRVEQLHPFGWKQMAANISQYPNLKEIIWC 947
Query 61 QEEPMNQGKERRINPII 77
QEEP+N G + P I
Sbjct 948 QEEPLNAGAWTYMEPRI 964
> Hs18375678
Length=919
Score = 54.7 bits (130), Expect = 4e-08, Method: Composition-based stats.
Identities = 24/75 (32%), Positives = 45/75 (60%), Gaps = 7/75 (9%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G+ +Y L+ +RE + ++ +D A+ RVE+L PFP D +++ +Y +++ +W+
Sbjct 810 GKHFYSLVKQRESLGAKKHD-------FAIIRVEELCPFPLDSLQQEMSKYKHVKDHIWS 862
Query 61 QEEPMNQGKERRINP 75
QEEP N G ++P
Sbjct 863 QEEPQNMGPWSFVSP 877
> YIL125w
Length=1014
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 27/75 (36%), Positives = 35/75 (46%), Gaps = 9/75 (12%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
GQ+Y L RE + + A ++EQL PFPF + YPNL +VW
Sbjct 898 GQVYTALHKRRESLGDKTT---------AFLKIEQLHPFPFAQLRDSLNSYPNLEEIVWC 948
Query 61 QEEPMNQGKERRINP 75
QEEP+N G P
Sbjct 949 QEEPLNMGSWAYTEP 963
> At3g55410
Length=1009
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/75 (37%), Positives = 43/75 (57%), Gaps = 18/75 (24%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G++YY+L ER+++ + + VA+ RVEQL PFP+DL R+++RYP
Sbjct 899 GKVYYELDDERKKVGATD---------VAICRVEQLCPFPYDLIQRELKRYP-------- 941
Query 61 QEEPMNQGKERRINP 75
+E MN G I+P
Sbjct 942 -KEAMNMGAFSYISP 955
> 7302023
Length=901
Score = 47.4 bits (111), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 26/75 (34%), Positives = 38/75 (50%), Gaps = 9/75 (12%)
Query 1 GQIYYDLLSERERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWA 60
G+ YY L ERE+ Q+ + A+ R+E L PFP + +Y N++S VW+
Sbjct 793 GKHYYTLAEEREKRQAYDT---------AILRLESLCPFPIQELQAQLAQYGNVQSFVWS 843
Query 61 QEEPMNQGKERRINP 75
QEE N G + P
Sbjct 844 QEEHRNMGAWTFVRP 858
> CE23468
Length=906
Score = 43.5 bits (101), Expect = 1e-04, Method: Composition-based stats.
Identities = 20/50 (40%), Positives = 28/50 (56%), Gaps = 0/50 (0%)
Query 26 NRVALARVEQLSPFPFDLAIRDIQRYPNLRSVVWAQEEPMNQGKERRINP 75
+ VA+ RVE L PFP +++YP + VW+QEEP N G + P
Sbjct 815 DSVAIVRVEMLCPFPVVDLQAVLKKYPGAQDFVWSQEEPRNAGAWSFVRP 864
> YGR032w
Length=1895
Score = 34.7 bits (78), Expect = 0.040, Method: Composition-based stats.
Identities = 16/57 (28%), Positives = 32/57 (56%), Gaps = 0/57 (0%)
Query 28 VALARVEQLSPFPFDLAIRDIQRYPNLRSVVWAQEEPMNQGKERRINPIIMDKEVDI 84
V++ R+ + P + A ++ YP+L+ +E P+N+G+E RI ++D +I
Sbjct 1021 VSMQRLAKFKPHELENAEFLLRAYPDLQIAYLDEEPPLNEGEEPRIYSALIDGHCEI 1077
> YLR342w
Length=1876
Score = 32.7 bits (73), Expect = 0.18, Method: Composition-based stats.
Identities = 15/57 (26%), Positives = 31/57 (54%), Gaps = 0/57 (0%)
Query 28 VALARVEQLSPFPFDLAIRDIQRYPNLRSVVWAQEEPMNQGKERRINPIIMDKEVDI 84
V++ R+ + P + A ++ YP+L+ +E P+ +G+E RI ++D +I
Sbjct 1002 VSMQRLAKFKPHELENAEFLLRAYPDLQIAYLDEEPPLTEGEEPRIYSALIDGHCEI 1058
> At2g17120
Length=350
Score = 30.4 bits (67), Expect = 0.76, Method: Composition-based stats.
Identities = 13/34 (38%), Positives = 22/34 (64%), Gaps = 3/34 (8%)
Query 45 IRDIQRY---PNLRSVVWAQEEPMNQGKERRINP 75
+R+IQ NLRS++ A P+N +++R+NP
Sbjct 52 LRNIQTLFAVKNLRSILGANNLPLNTSRDQRVNP 85
> 7298307
Length=440
Score = 30.4 bits (67), Expect = 0.84, Method: Composition-based stats.
Identities = 19/53 (35%), Positives = 28/53 (52%), Gaps = 7/53 (13%)
Query 12 ERIQSENNDPDNPGNRVALARVEQLSPFP-FDLAIRDIQ------RYPNLRSV 57
E I +DPD+ N+ A ARV+ L FDL+ D + +YP LR++
Sbjct 384 EGIWVSYDDPDSASNKAAYARVKNLGGVALFDLSYDDFRGQCSGDKYPILRAI 436
> At1g67140
Length=2149
Score = 27.7 bits (60), Expect = 4.9, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 12 ERIQSENNDPDNPGNRVALARVEQLSPFPFDLAIRDIQRYP 52
+ ++S+N + +N G + LA E SP DL + IQ+ P
Sbjct 2079 QSVESKNLESENIGTDIKLASTEVESPALDDLEPQQIQKSP 2119
Lambda K H
0.317 0.138 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1197406694
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40