bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0799_orf1
Length=151
Score E
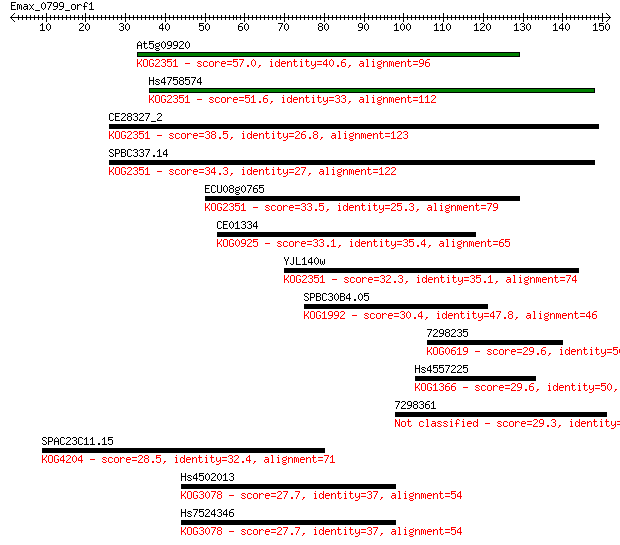
Sequences producing significant alignments: (Bits) Value
At5g09920 57.0 1e-08
Hs4758574 51.6 5e-07
CE28327_2 38.5 0.005
SPBC337.14 34.3 0.10
ECU08g0765 33.5 0.15
CE01334 33.1 0.20
YJL140w 32.3 0.35
SPBC30B4.05 30.4 1.2
7298235 29.6 2.0
Hs4557225 29.6 2.2
7298361 29.3 3.1
SPAC23C11.15 28.5 4.9
Hs4502013 27.7 9.1
Hs7524346 27.7 9.4
> At5g09920
Length=138
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 39/100 (39%), Positives = 55/100 (55%), Gaps = 5/100 (5%)
Query 33 LGPEFKNAKCLNLCELQLILG---DQLRLSVSRPA-EAHNLIKASYEYSSRFGKMTVRTA 88
+G EF AKCL CE+ LIL +QL+ P + + + S +Y RF + A
Sbjct 14 IGDEFLKAKCLMNCEVSLILEHKFEQLQQISEDPMNQVSQVFEKSLQYVKRFSRYKNPDA 73
Query 89 VVEIRKHLEREGDLQQFELALLVNLLPRTPDEAKAHIPSL 128
V ++R+ L R L +FEL +L NL P T +EA A +PSL
Sbjct 74 VRQVREILSRH-QLTEFELCVLGNLCPETVEEAVAMVPSL 112
> Hs4758574
Length=142
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 37/115 (32%), Positives = 57/115 (49%), Gaps = 4/115 (3%)
Query 36 EFKNAKCLNLCELQLILG--DQLRLSVSRPAEAHNLIKASYEYSSRFGKMTVRTAVVEIR 93
EF+ A+ L E+ ++L Q S E + + Y++RF + R + +R
Sbjct 25 EFETAETLLNSEVHMLLEHRKQQNESAEDEQELSEVFMKTLNYTARFSRFKNRETIASVR 84
Query 94 KHLEREGDLQQFELALLVNLLPRTPDEAKAHIPSLI-RLSPARLSRIIDTLEVFR 147
L + L +FELA L NL P T +E+KA IPSL R L +I+D ++ R
Sbjct 85 SLL-LQKKLHKFELACLANLCPETAEESKALIPSLEGRFEDEELQQILDDIQTKR 138
> CE28327_2
Length=133
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 33/128 (25%), Positives = 58/128 (45%), Gaps = 6/128 (4%)
Query 26 QDSLAGDLGPEFKNAKCLNL--CELQLILGDQLRLSVSRPA--EAHNLIKASYEYSSRFG 81
+D+ EF+ C L E+ L+L + + S S+ E + + Y+ R
Sbjct 5 EDAAECKFPKEFETTTCDALLTAEVYLLLEHRRQSSESKDEIEEMSEVFIKTLNYARRMS 64
Query 82 KMTVRTAVVEIRKHLEREGDLQQFELALLVNLLPRTPDEAKAHIPSL-IRLSPARLSRII 140
+ R + +R E L +FE+A + NL P +EAKA +PSL ++ + L ++
Sbjct 65 RFKNRETIRAVRAIF-SEKHLHKFEVAQIANLCPENAEEAKALVPSLENKIEESELEEVL 123
Query 141 DTLEVFRV 148
L+ R
Sbjct 124 KDLQSKRT 131
> SPBC337.14
Length=135
Score = 34.3 bits (77), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 33/129 (25%), Positives = 55/129 (42%), Gaps = 12/129 (9%)
Query 26 QDSLAGDLGPEFKNAKCLNLCELQLILGDQLRLSVSRPAEAH------NLIKASYEYSSR 79
+D+ LGPEF+N L + E ++++ L R E + +++K + Y +
Sbjct 8 EDAAQLKLGPEFENEDMLTVSEAKILIETVL---AQRARETNGEIPMTDVMKKTVAYFNV 64
Query 80 FGKMTVRTAVVEIRKHLEREGDLQQFELALLVNLLPRTPDEAKAHIPSLI-RLSPARLSR 138
F + A + L +FE A L L +EA+ IPSL ++ L
Sbjct 65 FARFKTAEATYACERILGNR--FHKFERAQLGTLCCEDAEEARTLIPSLANKIDDQNLQG 122
Query 139 IIDTLEVFR 147
I+D L R
Sbjct 123 ILDELSTLR 131
> ECU08g0765
Length=104
Score = 33.5 bits (75), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 20/79 (25%), Positives = 41/79 (51%), Gaps = 1/79 (1%)
Query 50 LILGDQLRLSVSRPAEAHNLIKASYEYSSRFGKMTVRTAVVEIRKHLEREGDLQQFELAL 109
L+ G + R + A + +++ Y F ++ ++ ++R L G + E+AL
Sbjct 8 LLEGQKERFRADFRSNASKVFRSTLGYLDDFCRIKDKSVAEDLRTTLSGLG-FGEVEIAL 66
Query 110 LVNLLPRTPDEAKAHIPSL 128
+L P++ +EAK+ +PSL
Sbjct 67 FGSLFPQSVEEAKSLVPSL 85
> CE01334
Length=739
Score = 33.1 bits (74), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 34/69 (49%), Gaps = 4/69 (5%)
Query 53 GDQLRLSVSRPAEAHNLIKASYEYSSRFGKMTVRTAVVE----IRKHLEREGDLQQFELA 108
D +R +SR + +NL + S ++ SR + +R A+V HLER G +
Sbjct 601 ADTVRTQLSRVMDKYNLRRVSTDFKSRDYYLNIRKALVAGFFMQVAHLERSGHYVTVKDN 660
Query 109 LLVNLLPRT 117
LVNL P T
Sbjct 661 QLVNLHPST 669
> YJL140w
Length=221
Score = 32.3 bits (72), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 26/75 (34%), Positives = 40/75 (53%), Gaps = 2/75 (2%)
Query 70 IKASYEYSSRFGKMTVRTAVVEIRKHLEREGDLQQFELALLVNLLPRTPDEAKAHIPSL- 128
+K + +Y + F + + V + + L+ G L FE+A L +L T DEAK IPSL
Sbjct 141 LKNTMQYLTNFSRFRDQETVGAVIQLLKSTG-LHPFEVAQLGSLACDTADEAKTLIPSLN 199
Query 129 IRLSPARLSRIIDTL 143
++S L RI+ L
Sbjct 200 NKISDDELERILKEL 214
> SPBC30B4.05
Length=967
Score = 30.4 bits (67), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 22/51 (43%), Positives = 29/51 (56%), Gaps = 5/51 (9%)
Query 75 EYSSRFGK-MTVRTAVVEIRKHLE---REGDLQQF-ELALLVNLLPRTPDE 120
+Y GK M T+V+ IRKH E ++ LQQF EL +L N+ R DE
Sbjct 300 KYDGLVGKAMAFLTSVIRIRKHAEFFQQDQVLQQFIELVVLPNICLRESDE 350
> 7298235
Length=1021
Score = 29.6 bits (65), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 17/36 (47%), Positives = 22/36 (61%), Gaps = 2/36 (5%)
Query 106 ELALLVNLLPRTPDEAKAHIPSLIR--LSPARLSRI 139
EL L N + R PD+A H+P L++ LS RLS I
Sbjct 188 ELRLSGNPILRVPDDAFGHVPQLVKLELSDCRLSHI 223
> Hs4557225
Length=1474
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 103 QQFELALLVNLLPRTPDEAKAHIPSLIRLS 132
++F AL V LP+T DE KAH I LS
Sbjct 1337 EEFPFALGVQTLPQTCDEPKAHTSFQISLS 1366
> 7298361
Length=1441
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 30/54 (55%), Gaps = 2/54 (3%)
Query 98 REGDLQQFELALLVNLLPRT-PDEAKAHIPSLIRLSPARLSRIIDTLEVFRVHA 150
++G + + ALL +L +T PD + P LI S +L ++ TL++ R HA
Sbjct 157 KKGGIMKGTRALLAHLSSQTRPDNVQGE-PILIETSEKQLEEVVITLDIGRAHA 209
> SPAC23C11.15
Length=1075
Score = 28.5 bits (62), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 36/75 (48%), Gaps = 5/75 (6%)
Query 9 SLHGVLGF----DRTKMDSPTQDSLAGDLGPEFKNAKCLNLCELQLILGDQLRLSVSRPA 64
+ H +GF R + +P Q D +FKN++C +L ELQ I+ L S
Sbjct 141 TYHRAIGFVSRVRRALLSNPEQFFKLQDSLRKFKNSEC-SLSELQTIVTSLLAEHPSLAH 199
Query 65 EAHNLIKASYEYSSR 79
E HN + +S + S+
Sbjct 200 EFHNFLPSSIFFGSK 214
> Hs4502013
Length=239
Score = 27.7 bits (60), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 27/55 (49%), Gaps = 5/55 (9%)
Query 44 NLCELQLILGDQLRLSVSRPAEAHNLIKASYEYSSRFGKMTVRTAVVE-IRKHLE 97
N C L GD LR V+ +E +KA+ + GK+ VVE I K+LE
Sbjct 38 NFCVCHLATGDMLRAMVASGSELGKKLKATMDA----GKLVSDEMVVELIEKNLE 88
> Hs7524346
Length=232
Score = 27.7 bits (60), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 27/55 (49%), Gaps = 5/55 (9%)
Query 44 NLCELQLILGDQLRLSVSRPAEAHNLIKASYEYSSRFGKMTVRTAVVE-IRKHLE 97
N C L GD LR V+ +E +KA+ + GK+ VVE I K+LE
Sbjct 38 NFCVCHLATGDMLRAMVASGSELGKKLKATMDA----GKLVSDEMVVELIEKNLE 88
Lambda K H
0.320 0.136 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1888713112
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40