bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0774_orf4
Length=231
Score E
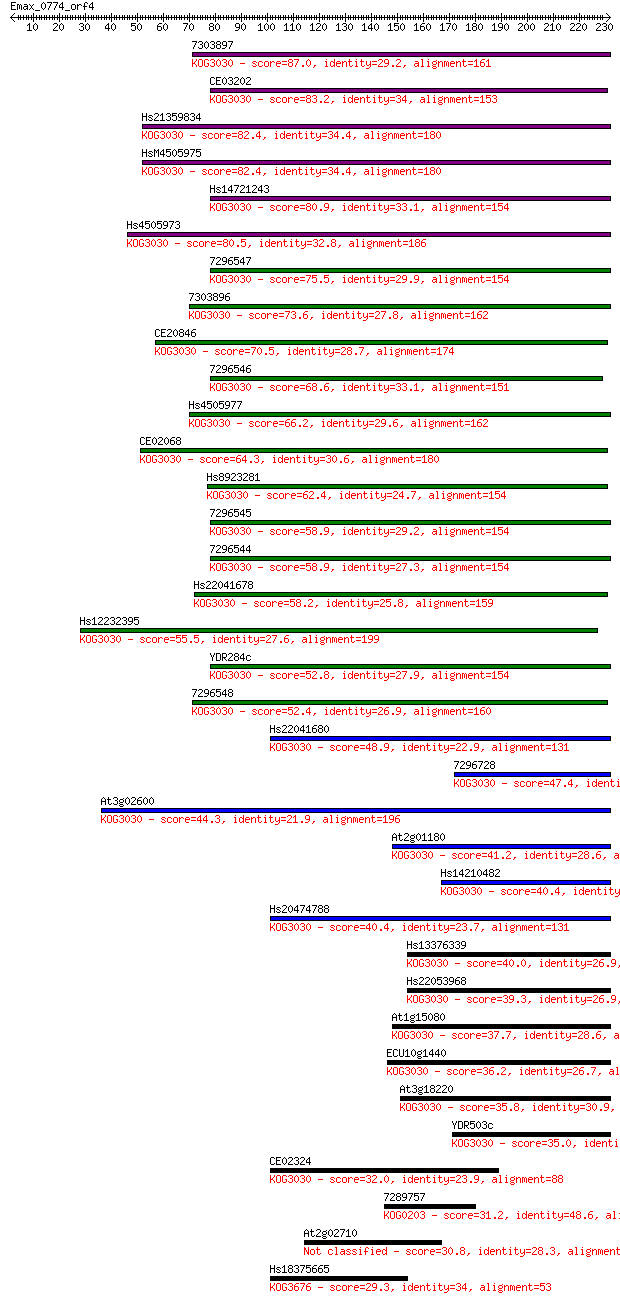
Sequences producing significant alignments: (Bits) Value
7303897 87.0 3e-17
CE03202 83.2 4e-16
Hs21359834 82.4 6e-16
HsM4505975 82.4 7e-16
Hs14721243 80.9 2e-15
Hs4505973 80.5 3e-15
7296547 75.5 9e-14
7303896 73.6 3e-13
CE20846 70.5 3e-12
7296546 68.6 9e-12
Hs4505977 66.2 6e-11
CE02068 64.3 2e-10
Hs8923281 62.4 8e-10
7296545 58.9 7e-09
7296544 58.9 9e-09
Hs22041678 58.2 1e-08
Hs12232395 55.5 1e-07
YDR284c 52.8 6e-07
7296548 52.4 7e-07
Hs22041680 48.9 1e-05
7296728 47.4 2e-05
At3g02600 44.3 2e-04
At2g01180 41.2 0.002
Hs14210482 40.4 0.003
Hs20474788 40.4 0.003
Hs13376339 40.0 0.004
Hs22053968 39.3 0.006
At1g15080 37.7 0.021
ECU10g1440 36.2 0.057
At3g18220 35.8 0.073
YDR503c 35.0 0.13
CE02324 32.0 1.1
7289757 31.2 2.0
At2g02710 30.8 2.7
Hs18375665 29.3 6.6
> 7303897
Length=372
Score = 87.0 bits (214), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 47/165 (28%), Positives = 81/165 (49%), Gaps = 4/165 (2%)
Query 71 LTGTATVRGSFCNNTDIALPKKKGSINLGQLMVLSFVVPSIIIILVELLIALVRATEDKE 130
L G RG FC++ + P ++ L + V+P +I +VE++I+ +A +D
Sbjct 102 LLGEPYKRGFFCDDESLKHPFHDSTVRNWMLYFIGAVIPVGVIFIVEVIISQNKAKQDNG 161
Query 131 AQQSSNVRMFCWEVPQWIVDLYTYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPD 190
S +E+P W+++ Y +G + F + L D K +G LRPHF C+P
Sbjct 162 NATSRRYVFMNYELPDWMIECYKKIGIYAFGAVLSQLTTDIAKYSIGRLRPHFIAVCQPQ 221
Query 191 WSQ-VKCKGSNGEYVYVEDFHCM---NDAHRVEDARRSFPSGHST 231
+ C + Y+++F C + A +++ R SFPSGHS+
Sbjct 222 MADGSTCDDAINAGKYIQEFTCKGVGSSARMLKEMRLSFPSGHSS 266
> CE03202
Length=318
Score = 83.2 bits (204), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 52/155 (33%), Positives = 76/155 (49%), Gaps = 18/155 (11%)
Query 78 RGSFCNNTDIALPKKKGSINLGQLMVLSFVVPSIIIILVELLIALVRATEDKEAQQSSNV 137
R C + I P K+ ++ L L+V++ P +I+ LVE ++ ++ A+ S
Sbjct 80 RAMPCGDISIQQPFKENTVGLKHLLVITLGSPFLIVALVEAILHFKSKGSNRLAKFFSAT 139
Query 138 RMFCWEVPQWIVDLYTYLGGFGFTMATAWLFA-DSLKCFVGSLRPHFFDACKPDWSQVKC 196
+ TYL M A FA + LKC+VG LRPHFF CKPDWS+V C
Sbjct 140 TI-------------TYLKYL--LMYAACTFAMEFLKCYVGRLRPHFFSVCKPDWSKVDC 184
Query 197 KGSNGEYVYVEDFHCMN-DAHRVEDARRSFPSGHS 230
++ D C N + ++ AR SFPSGH+
Sbjct 185 TDKQS-FIDSSDLVCTNPNPRKIRTARTSFPSGHT 218
> Hs21359834
Length=311
Score = 82.4 bits (202), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 62/188 (32%), Positives = 94/188 (50%), Gaps = 27/188 (14%)
Query 52 RVLMHILSLLAVCLIAANS---LTGTATV----RGSFCNNTDIALPKKKG-SINLGQLMV 103
RVL+ L L CL A + T+T+ RG +CN+ I P K G +IN L
Sbjct 33 RVLLICLDLF--CLFMAGLPFLIIETSTIKPYHRGFYCNDESIKYPLKTGETINDAVLCA 90
Query 104 LSFVVPSIIIILVELLIALVRATEDKEAQQSSNVRMFCWEVPQWIVDLYTYLGGFGFTMA 163
+ V+ + II E R K+++ + + P ++ LY +G F F A
Sbjct 91 VGIVIAILAIITGEF----YRIYYLKKSRST-------IQNP-YVAALYKQVGCFLFGCA 138
Query 164 TAWLFADSLKCFVGSLRPHFFDACKPDWSQVKCKGSNGEYVYVEDFHCMNDAHRVEDARR 223
+ F D K +G LRPHF C PD+SQ+ C S G Y++++ C D +V++AR+
Sbjct 139 ISQSFTDIAKVSIGRLRPHFLSVCNPDFSQINC--SEG---YIQNYRCRGDDSKVQEARK 193
Query 224 SFPSGHST 231
SF SGH++
Sbjct 194 SFFSGHAS 201
> HsM4505975
Length=311
Score = 82.4 bits (202), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 62/188 (32%), Positives = 94/188 (50%), Gaps = 27/188 (14%)
Query 52 RVLMHILSLLAVCLIAANS---LTGTATV----RGSFCNNTDIALPKKKG-SINLGQLMV 103
RVL+ L L CL A + T+T+ RG +CN+ I P K G +IN L
Sbjct 33 RVLLICLDLF--CLFMAGLPFLIIETSTIKPYHRGFYCNDESIKYPLKTGETINDAVLCA 90
Query 104 LSFVVPSIIIILVELLIALVRATEDKEAQQSSNVRMFCWEVPQWIVDLYTYLGGFGFTMA 163
+ V+ + II E R K+++ + + P ++ LY +G F F A
Sbjct 91 VGIVIAILAIITGEF----YRIYYLKKSRST-------IQNP-YVAALYKQVGCFLFGCA 138
Query 164 TAWLFADSLKCFVGSLRPHFFDACKPDWSQVKCKGSNGEYVYVEDFHCMNDAHRVEDARR 223
+ F D K +G LRPHF C PD+SQ+ C S G Y++++ C D +V++AR+
Sbjct 139 ISQSFTDIAKVSIGRLRPHFLSVCNPDFSQINC--SEG---YIQNYRCRGDDSKVQEARK 193
Query 224 SFPSGHST 231
SF SGH++
Sbjct 194 SFFSGHAS 201
> Hs14721243
Length=285
Score = 80.9 bits (198), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 51/154 (33%), Positives = 74/154 (48%), Gaps = 16/154 (10%)
Query 78 RGSFCNNTDIALPKKKGSINLGQLMVLSFVVPSIIIILVELLIALVRATEDKEAQQSSNV 137
RG FC + I P ++ L+++ +P IIL E L +S +
Sbjct 37 RGFFCKDNSINYPYHDSTVTSTVLILVGVGLPISSIILGETLSVYCNLL-----HSNSFI 91
Query 138 RMFCWEVPQWIVDLYTYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSQVKCK 197
R +I +Y +G F F A + D K +G LRPHF D C PDWS++ C
Sbjct 92 R------NNYIATIYKAIGTFLFGAAASQSLTDIAKYSIGRLRPHFLDVCDPDWSKINC- 144
Query 198 GSNGEYVYVEDFHCMNDAHRVEDARRSFPSGHST 231
S+G Y+E + C +A RV++ R SF SGHS+
Sbjct 145 -SDG---YIEYYICRGNAERVKEGRLSFYSGHSS 174
> Hs4505973
Length=289
Score = 80.5 bits (197), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 61/188 (32%), Positives = 89/188 (47%), Gaps = 18/188 (9%)
Query 46 IPSYPIRVLMHILSLLAVCLIAANSLTGTAT--VRGSFCNNTDIALPKKKGSINLGQLMV 103
+P + VL +L+ L + + +T T RG FCN+ I P K+ +I L
Sbjct 7 LPYVALDVLCVLLAGLPFAIFTSRHITSRHTPFQRGVFCNDESIKYPYKEDTIPYALLGG 66
Query 104 LSFVVPSIIIILVELLIALVRATEDKEAQQSSNVRMFCWEVPQWIVDLYTYLGGFGFTMA 163
+ I+IIL E L +S +R +I +Y +G F F A
Sbjct 67 IIIPFSIIVIILGETLSVYCNLL-----HSNSFIR------NNYIATIYKAIGTFLFGAA 115
Query 164 TAWLFADSLKCFVGSLRPHFFDACKPDWSQVKCKGSNGEYVYVEDFHCMNDAHRVEDARR 223
+ D K +G LRPHF D C PDWS++ C S+G Y+E + C +A RV++ R
Sbjct 116 ASQSLTDIAKYSIGRLRPHFLDVCDPDWSKINC--SDG---YIEYYICRGNAERVKEGRL 170
Query 224 SFPSGHST 231
SF SGHS+
Sbjct 171 SFYSGHSS 178
> 7296547
Length=340
Score = 75.5 bits (184), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 46/158 (29%), Positives = 78/158 (49%), Gaps = 10/158 (6%)
Query 78 RGSFCNNTDIALPKKKGSINLGQLMVLSFVVPSIIIILVELLIALVRATEDKEAQQSSNV 137
RG FC++ I+ P + +I L ++ ++P++++++VE + L + S+ V
Sbjct 64 RGFFCDDESISYPFQDNTITPVMLGLIVGLLPALVMVVVEYVSHL------RAGDISATV 117
Query 138 RMFCWEVPQWIVDLYTYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSQ-VKC 196
+ W V W V+L F F + + + K +G LRPHF C+P + C
Sbjct 118 DLLGWRVSTWYVELGRQSTYFCFGLLLTFDATEVGKYTIGRLRPHFLAVCQPQIADGSMC 177
Query 197 KGSNGEYVYVEDFHCMNDAHRVED---ARRSFPSGHST 231
+ Y+E++ C + VED AR SFPSGHS+
Sbjct 178 SDPVNLHRYMENYDCAGEGFTVEDVRQARLSFPSGHSS 215
> 7303896
Length=246
Score = 73.6 bits (179), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 45/168 (26%), Positives = 80/168 (47%), Gaps = 6/168 (3%)
Query 70 SLTGTATVRGSFCNNTDIALPKKKGSINLGQLMVLSFVVPSIIIILVELLIALVRATEDK 129
SL G A RG FC+++ + P + ++ L ++ +P ++++VE +
Sbjct 5 SLWGEAVKRGFFCDDSSLRHPYRDSTMPSWILYLMCGALPLTVMLVVEFFRGQDKRLHSP 64
Query 130 --EAQQSSNVRMFCWEVPQWIVDLYTYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDAC 187
++ S + E+P W+V+ Y +G F F + L + K +G LRPHF+ C
Sbjct 65 FPKSTMCSGYHLCHLELPTWLVECYHRMGIFIFGLGVEQLSTNIAKYSIGRLRPHFYTLC 124
Query 188 KPDWSQ-VKCKGSNGEYVYVEDFHCMN---DAHRVEDARRSFPSGHST 231
+P C Y+E+F C + +++D R SFPSGH++
Sbjct 125 QPVMKDGTTCSDPINAARYIEEFTCAAVDITSKQLKDMRLSFPSGHAS 172
> CE20846
Length=346
Score = 70.5 bits (171), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 50/175 (28%), Positives = 83/175 (47%), Gaps = 7/175 (4%)
Query 57 ILSLLAVCLIAANSLTGTATVRGSFCNNTDIALPKKKGSINLGQLMVLSFVVPSIIIILV 116
+L+L+A+ L + RG +C++ I P + + L+V+ ++P ++I+
Sbjct 18 VLTLIAIPLYVFHEFIPPVR-RGFYCDDESIRYPFRDSKVTRQMLIVVGLLIPILLILAT 76
Query 117 ELLIALVRATEDKEAQQSSNVRMFCWEVPQWIVDLYTYLGGFGFTMATAWLFADSLKCFV 176
EL L A E K + + V + +V LY ++G F + L D K +
Sbjct 77 ELFRTL--AWEKKCETEFKTYHVRNHSVHRLVVRLYCFIGYFFVGVCFNQLMVDIAKYTI 134
Query 177 GSLRPHFFDACKPDWSQVKCKGSNGEYVYVEDFHC-MNDAHRVEDARRSFPSGHS 230
G RPHF D C+PD C + +Y+ DF C D ++ +A+ SF SGHS
Sbjct 135 GRQRPHFMDVCRPDIGYQTCSQPD---LYITDFKCTTTDTKKIHEAQLSFYSGHS 186
> 7296546
Length=305
Score = 68.6 bits (166), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 50/166 (30%), Positives = 78/166 (46%), Gaps = 25/166 (15%)
Query 78 RGSFCNNTDIALPKKKGSINLGQLMVLSFVVPSIIIILVELLIALVRATEDKEAQQSSNV 137
RG C++T + P ++ + L + +P+ +++VE+L A V + + Q+ V
Sbjct 40 RGFHCSDTSLKYPYRQPWLTKVHLTIAVVALPAAFVLVVEMLRAAVVPSSTELTQRFVFV 99
Query 138 RMFCWEVPQWIVDLY----TYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDW-- 191
+ +P++I + Y YL G G T+A L S G LRP+FFD C+P W
Sbjct 100 GV---RIPRFISECYKAIGVYLFGLGLTLAAIRLTKHS----TGRLRPYFFDICQPTWGT 152
Query 192 ------SQVKCKGSNGEYVYVEDFHCMNDAHR---VEDARRSFPSG 228
S V + S +Y+EDF C A + R SFPSG
Sbjct 153 EGGESCSDVTAQNST---LYLEDFSCTEFAASQDLLALVRHSFPSG 195
> Hs4505977
Length=288
Score = 66.2 bits (160), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 48/162 (29%), Positives = 73/162 (45%), Gaps = 17/162 (10%)
Query 70 SLTGTATVRGSFCNNTDIALPKKKGSINLGQLMVLSFVVPSIIIILVELLIALVRATEDK 129
+L RG +C + I P + +I G +++ V + +ILV A + T+
Sbjct 26 TLVNAPYKRGFYCGDDSIRYPYRPDTITHG---LMAGVTITATVILVSAGEAYLVYTDRL 82
Query 130 EAQQSSNVRMFCWEVPQWIVDLYTYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKP 189
++ N ++ +Y LG F F A + D K +G LRP+F C P
Sbjct 83 YSRSDFN---------NYVAAVYKVLGTFLFGAAVSQSLTDLAKYMIGRLRPNFLAVCDP 133
Query 190 DWSQVKCKGSNGEYVYVEDFHCMNDAHRVEDARRSFPSGHST 231
DWS+V C YV +E C + V +AR SF SGHS+
Sbjct 134 DWSRVNC----SVYVQLEKV-CRGNPADVTEARLSFYSGHSS 170
> CE02068
Length=341
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 55/185 (29%), Positives = 84/185 (45%), Gaps = 19/185 (10%)
Query 51 IRVLMHILSLLAVCLIAANSLTGTATVRGSFCNNTDIALPKKKGSINLGQLMVLSFVVPS 110
I + + L+ AV +I +L G + RG FC++ I +K +I QLM+ + V+ +
Sbjct 28 ISLFIFFLATAAVTVIVP-TLLGVSQ-RGFFCDDDSIRYEYRKDTITAVQLMLYNLVLNA 85
Query 111 IIIILVELLIALVRATEDKEAQQSSNVRMFCW---EVPQWIVDLYTYLG--GFGFTMATA 165
++ VE ++ + + N + W + V L TY G GF M A
Sbjct 86 ATVLFVEYY-------RMQKVESNINNPRYRWRNNHLHVLFVRLLTYFGYSQIGFVMNIA 138
Query 166 WLFADSLKCFVGSLRPHFFDACKPDWSQVKCKGSNGEYVYVEDFHCMNDAHRVEDARRSF 225
K VG LRPHF D CK + C + + Y+ D+ C V +AR+SF
Sbjct 139 LNIVT--KHVVGRLRPHFLDVCK--LANDTCVTGDS-HRYITDYTCTGPPELVLEARKSF 193
Query 226 PSGHS 230
SGHS
Sbjct 194 YSGHS 198
> Hs8923281
Length=325
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 38/160 (23%), Positives = 78/160 (48%), Gaps = 9/160 (5%)
Query 77 VRGSFCNNTDIALP----KKKGSINLGQLMVLSFVVPSIIIILVELLIALVRAT-EDKEA 131
++G FC + D+ P +++ I L + P+ II + E+ + +++T E A
Sbjct 45 IQGFFCQDGDLMKPYPGTEEESFITPLVLYCVLAATPTAIIFIGEISMYFIKSTRESLIA 104
Query 132 QQSSNVRMFCWEVPQWIVDLYTYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDW 191
++ + + C + + + + G F F + +F ++ + G L P+F CKP++
Sbjct 105 REKTILTGECCYLNPLLRRIIRFTGVFAFGLFATDIFVNAGQVVTGHLTPYFLTVCKPNY 164
Query 192 SQVKCKGSNGEYVYVEDFH-CMNDAHRVEDARRSFPSGHS 230
+ C+ + ++ + + C D +E ARRSFPS H+
Sbjct 165 TSADCQ---AHHQFINNGNICTGDLEVIEKARRSFPSKHA 201
> 7296545
Length=341
Score = 58.9 bits (141), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 45/164 (27%), Positives = 76/164 (46%), Gaps = 21/164 (12%)
Query 78 RGSFCNNTDIALPKKKGSINLGQLMVLSFVVPSIIIILVELLIALVRATEDKEAQQSSNV 137
RG FC + ++ P + G+I+ ++ + VP+ +I++VEL R +++
Sbjct 36 RGFFCGDETLSYPARDGTISSKVIIAIVLGVPNAVIVVVELF----RQLPGGPLREAGGK 91
Query 138 RMFCWEVPQWIVDLYT----YLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACK---PD 190
R C + + LY YL G T L K +G LRPHF C+ PD
Sbjct 92 RDSC-RIAHRLGVLYRQVIFYLYGLAMVTFTTML----TKLCLGRLRPHFLAVCQPMLPD 146
Query 191 WSQVKCKGSNGEYVYVEDFHCMN---DAHRVEDARRSFPSGHST 231
S C+ + Y++ F C N ++ ++ +SFPSGH++
Sbjct 147 GS--SCQDAQNLGRYIDSFTCSNANMTDYQFKELYQSFPSGHAS 188
> 7296544
Length=334
Score = 58.9 bits (141), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 42/158 (26%), Positives = 69/158 (43%), Gaps = 21/158 (13%)
Query 78 RGSFCNNTDIALPKKKGSINLGQLMVLSFVVPSIIIILVELLIALVRATEDKEAQQSSNV 137
RG FC++ I P K +I + L+++ ++P + + +VE++ R
Sbjct 6 RGFFCSDLSIRYPYKDCTITVPMLLLMMLLLPMLFVAVVEIMRICKR------------- 52
Query 138 RMFCWEVPQWIVDLYTYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSQ-VKC 196
+ + +L+ F F +L + K VG LRPHFF C+P C
Sbjct 53 ----FRTRLYFRNLWRAEATFSFGFIATYLTTELAKHAVGRLRPHFFHGCQPRLDDGSSC 108
Query 197 KGSNGEYVYVEDFHCMND---AHRVEDARRSFPSGHST 231
+YVE FHC N+ ++ + SFPS HS+
Sbjct 109 SDLQNAELYVEQFHCTNNNLSTRQIRELHVSFPSAHSS 146
> Hs22041678
Length=321
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 41/164 (25%), Positives = 80/164 (48%), Gaps = 7/164 (4%)
Query 72 TGTATVRGSFCNNTDIALP----KKKGSINLGQLMVLSFVVPSIIIILVELLI-ALVRAT 126
T T V+G FC+++ P + ++ L L+ VP ++II+ E + L AT
Sbjct 35 TFTVNVQGFFCHDSAYRKPYPGPEDSSAVPPVLLYSLAAGVPVLVIIVGETAVFCLQLAT 94
Query 127 EDKEAQQSSNVRMFCWEVPQWIVDLYTYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDA 186
D E Q+ + + C + + +LG + F + +F ++ + G+L PHF
Sbjct 95 RDFENQEKTILTGDCCYINPLVRRTVRFLGIYTFGLFATDIFVNAGQVVTGNLAPHFLAL 154
Query 187 CKPDWSQVKCKGSNGEYVYVEDFHCMNDAHRVEDARRSFPSGHS 230
CKP+++ + C+ +++ E+ C + + AR++FPS +
Sbjct 155 CKPNYTALGCQQYT-QFISGEE-ACTGNPDLIMRARKTFPSKEA 196
> Hs12232395
Length=343
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 55/212 (25%), Positives = 92/212 (43%), Gaps = 20/212 (9%)
Query 28 AGVPSSLKRSLGFSLGRVIPSYPI--RVLMHILSLLAVCLIAANSLTGTATVRGSFCNNT 85
AG LKRS +IP + VL+ I+ LLA L ++L +G FC ++
Sbjct 2 AGGRPHLKRSFS-----IIPCFVFVESVLLGIVILLAYRLEFTDTLP--VHTQGFFCYDS 54
Query 86 DIA--LPKKKGSINLGQLMVLSFVV--PSIIIILVELLIALVRATEDKEA--QQSSNVRM 139
A P + + + +V + V P++ I+L EL A A +S+ V
Sbjct 55 TYAKPYPGPEAASRVPPALVYALVTAGPTLTILLGELARAFFPAPPSAVPVIGESTIVSG 114
Query 140 FCWEVPQWIVDLYTYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSQVKCKGS 199
C + L +LG + F + T +FA++ + G+ PHF C+P+++ + C
Sbjct 115 ACCRFSPPVRRLVRFLGVYSFGLFTTTIFANAGQVVTGNPTPHFLSVCRPNYTALGCLPP 174
Query 200 NGEYVYVEDF-----HCMNDAHRVEDARRSFP 226
+ + + F C V ARR+FP
Sbjct 175 SPDRPGPDRFVTDQGACAGSPSLVAAARRAFP 206
> YDR284c
Length=289
Score = 52.8 bits (125), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 43/156 (27%), Positives = 66/156 (42%), Gaps = 29/156 (18%)
Query 78 RGSFCNNTDIALPKKKGS-INLGQLMVLSFVVPSIIIILVELLIALVRATEDKEAQQSSN 136
R + N+ I+ P +N L V SFVVPS+ I+++ ++A R
Sbjct 43 RQFYINDLTISHPYATTERVNNNMLFVYSFVVPSLTILIIGSILADRR------------ 90
Query 137 VRMFCWEVPQWIVDLYTYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSQVKC 196
I LYT L G + F + +K ++G LRP F D C+P
Sbjct 91 ---------HLIFILYTSLLGLSLAWFSTSFFTNFIKNWIGRLRPDFLDRCQP------V 135
Query 197 KGSNGEYVYVEDFHCMNDAH-RVEDARRSFPSGHST 231
+G + ++ C H R+ D R+ PSGHS+
Sbjct 136 EGLPLDTLFTAKDVCTTKNHERLLDGFRTTPSGHSS 171
> 7296548
Length=305
Score = 52.4 bits (124), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 43/168 (25%), Positives = 67/168 (39%), Gaps = 24/168 (14%)
Query 71 LTGTATVRGSFCNNTDIALPKKKGSINLGQLMVLSFVVPSIIIILVELLIALVRATEDKE 130
L G T RG FC++ + P + +++ L L +P I ++++E +
Sbjct 33 LWGPPTKRGFFCDDESLMYPYHENTVSPTLLHWLGLYLPLISLVVLESFL---------- 82
Query 131 AQQSSNVRMFCWEVPQWIVDLYTYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACK-- 188
S M W + +Y + F + + L K +G LRPHFF C
Sbjct 83 ---SHRKDMAPWPT---LWPVYNTVRWFLYGYVSNDLLKGIGKQALGRLRPHFFAVCSPH 136
Query 189 -PDWSQVKCKGSNGEYVYVEDFHCMNDAHR-----VEDARRSFPSGHS 230
PD S + G Y D+ C + + + D SFPSGHS
Sbjct 137 FPDGSSCLDESHRGALKYHTDYECRPNLSQATEEMIRDVNVSFPSGHS 184
> Hs22041680
Length=763
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 30/131 (22%), Positives = 57/131 (43%), Gaps = 1/131 (0%)
Query 101 LMVLSFVVPSIIIILVELLIALVRATEDKEAQQSSNVRMFCWEVPQWIVDLYTYLGGFGF 160
L+ L+F P+I I++ E ++ + N+ ++ ++G F
Sbjct 125 LLSLAFAGPAITIMVGEGILYCCLSKRRNGVGLEPNINAGGCNFNSFLRRAVRFVGVHVF 184
Query 161 TMATAWLFADSLKCFVGSLRPHFFDACKPDWSQVKCKGSNGEYVYVEDFHCMNDAHRVED 220
+ + L D ++ G P+F CKP+++ + Y+ VED +D +
Sbjct 185 GLCSTALITDIIQLSTGYQAPYFLTVCKPNYTSLNVSCKENSYI-VEDICSGSDLTVINS 243
Query 221 ARRSFPSGHST 231
R+SFPS H+T
Sbjct 244 GRKSFPSQHAT 254
> 7296728
Length=412
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/60 (41%), Positives = 32/60 (53%), Gaps = 0/60 (0%)
Query 172 LKCFVGSLRPHFFDACKPDWSQVKCKGSNGEYVYVEDFHCMNDAHRVEDARRSFPSGHST 231
LK VG RP +F C PD V SNG + DF+C + + R+SFPSGHS+
Sbjct 215 LKITVGRPRPDYFYRCFPDGVMVLNTTSNGVDTSILDFNCTGLPGDINEGRKSFPSGHSS 274
> At3g02600
Length=314
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 43/197 (21%), Positives = 79/197 (40%), Gaps = 41/197 (20%)
Query 36 RSLGFSLGRV-IPSYPIRVLMHILSLLAVCLIAANSLTGTATVRGSFCNNTDIALPKKKG 94
RS G ++ R + + I VL+ IL + + + G + TD++ P K
Sbjct 12 RSHGMTVARTHMHDWIILVLLVILECVLLIIHPFYRFVGKDMM-------TDLSYPLKSN 64
Query 95 SINLGQLMVLSFVVPSIIIILVELLIALVRATEDKEAQQSSNVRMFCWEVPQWIVDLYTY 154
++ + + V + ++P +I I + R + + DL+
Sbjct 65 TVPIWSVPVYAMLLPLVIFIFIYF-----RRRD--------------------VYDLHHA 99
Query 155 LGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSQVKCKGSNGEYVYVEDFHCMND 214
+ G +++ + D++K VG RP FF C PD Y + D C D
Sbjct 100 VLGLLYSVLVTAVLTDAIKNAVGRPRPDFFWRCFPD--------GKALYDSLGDVICHGD 151
Query 215 AHRVEDARRSFPSGHST 231
+ + +SFPSGH++
Sbjct 152 KSVIREGHKSFPSGHTS 168
> At2g01180
Length=302
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 40/84 (47%), Gaps = 8/84 (9%)
Query 148 IVDLYTYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSQVKCKGSNGEYVYVE 207
+ DL+ + G F + + DS+K G RP+F+ C PD ++ Y +
Sbjct 93 VYDLHHSILGLLFAVLITGVITDSIKVATGRPRPNFYWRCFPDGKEL--------YDALG 144
Query 208 DFHCMNDAHRVEDARRSFPSGHST 231
C A V++ +SFPSGH++
Sbjct 145 GVVCHGKAAEVKEGHKSFPSGHTS 168
> Hs14210482
Length=175
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 31/65 (47%), Gaps = 12/65 (18%)
Query 167 LFADSLKCFVGSLRPHFFDACKPDWSQVKCKGSNGEYVYVEDFHCMNDAHRVEDARRSFP 226
+F +++K VG RP FF C PD + D C D V + R+SFP
Sbjct 61 VFTNTIKLIVGRPRPDFFYRCFPDG------------LAHSDLMCTGDKDVVNEGRKSFP 108
Query 227 SGHST 231
SGHS+
Sbjct 109 SGHSS 113
> Hs20474788
Length=271
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 31/131 (23%), Positives = 52/131 (39%), Gaps = 36/131 (27%)
Query 101 LMVLSFVVPSIIIILVELLIALVRATEDKEAQQSSNVRMFCWEVPQWIVDLYTYLGGFGF 160
+ +SF+ P +I +V++ +R T+ E +++
Sbjct 54 MFAISFLTPLAVICVVKI----IRRTDKTEIKEA--------------------FLAVSL 89
Query 161 TMATAWLFADSLKCFVGSLRPHFFDACKPDWSQVKCKGSNGEYVYVEDFHCMNDAHRVED 220
+A + +++K VG RP FF C PD V + HC D V +
Sbjct 90 ALALNGVCTNTIKLIVGRPRPDFFYRCFPDG------------VMNSEMHCTGDPDLVSE 137
Query 221 ARRSFPSGHST 231
R+SFPS HS+
Sbjct 138 GRKSFPSIHSS 148
> Hs13376339
Length=746
Score = 40.0 bits (92), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 21/78 (26%), Positives = 37/78 (47%), Gaps = 1/78 (1%)
Query 154 YLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSQVKCKGSNGEYVYVEDFHCMN 213
++G F + L D ++ G P F CKP+++ + Y+ +D +
Sbjct 133 FVGVHVFGLCATALVTDVIQLATGYHTPFFLTVCKPNYTLLGTSCEVNPYI-TQDICSGH 191
Query 214 DAHRVEDARRSFPSGHST 231
D H + AR++FPS H+T
Sbjct 192 DIHAILSARKTFPSQHAT 209
> Hs22053968
Length=686
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 21/78 (26%), Positives = 37/78 (47%), Gaps = 1/78 (1%)
Query 154 YLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSQVKCKGSNGEYVYVEDFHCMN 213
++G F + L D ++ G P F CKP+++ + Y+ +D +
Sbjct 73 FVGVHVFGLCATALVTDVIQLATGYHTPFFLTVCKPNYTLLGTSCEVNPYI-TQDICSGH 131
Query 214 DAHRVEDARRSFPSGHST 231
D H + AR++FPS H+T
Sbjct 132 DIHAILSARKTFPSQHAT 149
> At1g15080
Length=290
Score = 37.7 bits (86), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 40/84 (47%), Gaps = 7/84 (8%)
Query 148 IVDLYTYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSQVKCKGSNGEYVYVE 207
+ DL+ + G F++ + D++K VG RP FF C PD G + +
Sbjct 93 VYDLHHAILGLLFSVLITGVITDAIKDAVGRPRPDFFWRCFPD-------GIGIFHNVTK 145
Query 208 DFHCMNDAHRVEDARRSFPSGHST 231
+ C V++ +SFPSGH++
Sbjct 146 NVLCTGAKDVVKEGHKSFPSGHTS 169
> ECU10g1440
Length=234
Score = 36.2 bits (82), Expect = 0.057, Method: Compositional matrix adjust.
Identities = 23/86 (26%), Positives = 34/86 (39%), Gaps = 18/86 (20%)
Query 146 QWIVDLYTYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSQVKCKGSNGEYVY 205
+ I ++Y Y F T + ++ K G LRP F C P +
Sbjct 75 ERIHEIYFY-ASFLVTCLVGFAVVENTKNLAGRLRPDFLSRCNPVAGK------------ 121
Query 206 VEDFHCMNDAHRVEDARRSFPSGHST 231
C + V D R+SFPSGH++
Sbjct 122 -----CTGNPLVVLDGRKSFPSGHTS 142
> At3g18220
Length=301
Score = 35.8 bits (81), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 38/81 (46%), Gaps = 14/81 (17%)
Query 151 LYTYLGGFGFTMATAWLFADSLKCFVGSLRPHFFDACKPDWSQVKCKGSNGEYVYVEDFH 210
L+ + G GF+ + DS+K VG RP+FF C P NG+ D
Sbjct 96 LHHAILGIGFSCLVTGVTTDSIKDAVGRPRPNFFYRCFP----------NGK----PDVV 141
Query 211 CMNDAHRVEDARRSFPSGHST 231
C +++ +SFPSGH++
Sbjct 142 CHGVKKIIKEGYKSFPSGHTS 162
> YDR503c
Length=274
Score = 35.0 bits (79), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 31/61 (50%), Gaps = 3/61 (4%)
Query 171 SLKCFVGSLRPHFFDACKPDWSQVKCKGSNGEYVYVEDFHCMNDAHRVEDARRSFPSGHS 230
+LK +G+LRP F D C PD ++ + V+ D + + + +S PSGHS
Sbjct 134 ALKLIIGNLRPDFVDRCIPDLQKM---SDSDSLVFGLDICKQTNKWILYEGLKSTPSGHS 190
Query 231 T 231
+
Sbjct 191 S 191
> CE02324
Length=396
Score = 32.0 bits (71), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 21/91 (23%), Positives = 41/91 (45%), Gaps = 10/91 (10%)
Query 101 LMVLSFVVPSIIIILVELLIALVRATEDKEAQQSSN---VRMFCWEVPQWIVDLYTYLGG 157
L L+F +P ++I++ E++ L K + V +F + ++++ YL G
Sbjct 97 LYTLAFTIPPLVILIGEVMFWLFSTKPRKIVYANCGECPVHLFTRRLFRFVI---IYLAG 153
Query 158 FGFTMATAWLFADSLKCFVGSLRPHFFDACK 188
+ +F D++K G RP+F C
Sbjct 154 ----LLIVQIFVDTIKLMTGYQRPYFLSLCN 180
> 7289757
Length=1009
Score = 31.2 bits (69), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 17/35 (48%), Positives = 19/35 (54%), Gaps = 5/35 (14%)
Query 145 PQWIVDLYTYLGGFGFTMATAWLFADSLKCFVGSL 179
P+WIV L T GGF L+A S CFVG L
Sbjct 76 PKWIVFLKTMFGGFAI-----LLWAGSFLCFVGYL 105
> At2g02710
Length=429
Score = 30.8 bits (68), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 15/57 (26%), Positives = 29/57 (50%), Gaps = 4/57 (7%)
Query 114 ILVELLIALVRATEDKEAQQSSNVRMFCWEVPQW----IVDLYTYLGGFGFTMATAW 166
I +E ++ ++R TED S N +++ W++ I +LY+ G+G + W
Sbjct 363 ITIENVLLVIRQTEDISNLYSKNTKIYRWKMQHRKVEDISNLYSKTVGYGHRILHLW 419
> Hs18375665
Length=839
Score = 29.3 bits (64), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 35/53 (66%), Gaps = 5/53 (9%)
Query 101 LMVLSFVVPSIIIILVELLIALVRATEDKEAQQSSNVRMFCWEVPQWIVDLYT 153
+++L++V+ + I++L +LIAL+ T +K AQ+S N+ W++ + I L T
Sbjct 662 ILLLAYVILTYILLL-NMLIALMGETVNKIAQESKNI----WKLQRAITILDT 709
Lambda K H
0.323 0.136 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4491129636
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40