bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
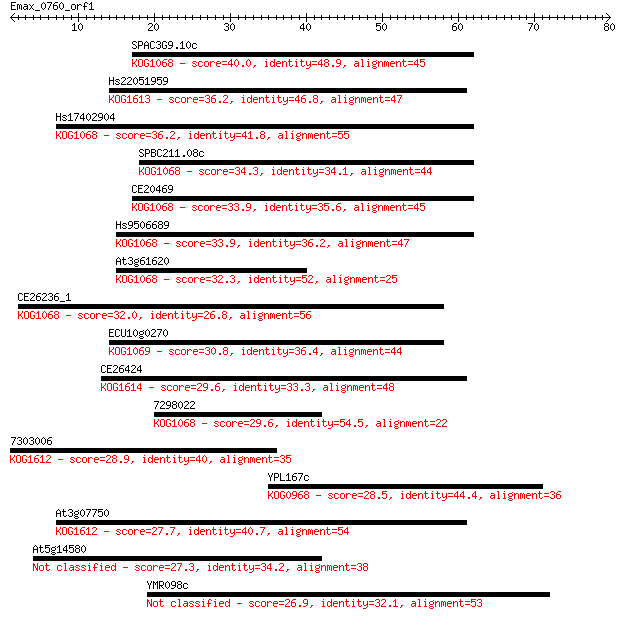
Query= Emax_0760_orf1
Length=79
Score E
Sequences producing significant alignments: (Bits) Value
SPAC3G9.10c 40.0 0.001
Hs22051959 36.2 0.014
Hs17402904 36.2 0.015
SPBC211.08c 34.3 0.064
CE20469 33.9 0.068
Hs9506689 33.9 0.076
At3g61620 32.3 0.24
CE26236_1 32.0 0.30
ECU10g0270 30.8 0.56
CE26424 29.6 1.4
7298022 29.6 1.4
7303006 28.9 2.5
YPL167c 28.5 3.1
At3g07750 27.7 5.3
At5g14580 27.3 6.6
YMR098c 26.9 8.0
> SPAC3G9.10c
Length=242
Score = 40.0 bits (92), Expect = 0.001, Method: Composition-based stats.
Identities = 22/47 (46%), Positives = 29/47 (61%), Gaps = 2/47 (4%)
Query 17 SASGSAFVSLGNTKVYCSVFGP-RPAGRSDLQ-DRGFIKVDFRSSPF 61
S +GSAF+ LGNTKV C V GP P +S + DR F+ V+ + F
Sbjct 34 SENGSAFIELGNTKVLCIVDGPSEPVIKSKARADRTFVNVEINIASF 80
> Hs22051959
Length=276
Score = 36.2 bits (82), Expect = 0.014, Method: Composition-based stats.
Identities = 22/57 (38%), Positives = 29/57 (50%), Gaps = 10/57 (17%)
Query 14 ALGSASGSAFVSLGNTKVYCSVFGPRPAGRSDLQDRGFI--KVD--------FRSSP 60
++ +A GSA V LGNT V C V A +D D+G++ VD FRS P
Sbjct 41 SISTADGSALVKLGNTTVICGVKAEFAAPSTDAPDKGYVVPNVDLPPLCSSRFRSGP 97
> Hs17402904
Length=272
Score = 36.2 bits (82), Expect = 0.015, Method: Composition-based stats.
Identities = 23/68 (33%), Positives = 32/68 (47%), Gaps = 13/68 (19%)
Query 7 PMHLQTLALGSASGSAFVSLGNTKVYCSVFGPRPAGRSDLQD-------------RGFIK 53
P++ + L A GSA++ G TKV C+V GPR A + RG +
Sbjct 38 PVYARAGLLSQAKGSAYLEAGGTKVLCAVSGPRQAEGGERGGGPAGAGGEAPAALRGRLL 97
Query 54 VDFRSSPF 61
DFR +PF
Sbjct 98 CDFRRAPF 105
> SPBC211.08c
Length=257
Score = 34.3 bits (77), Expect = 0.064, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 18 ASGSAFVSLGNTKVYCSVFGPRPAGRSDLQDRGFIKVDFRSSPF 61
A GS++ K+ C+V GPRP+ + + +FR SPF
Sbjct 48 AVGSSYFESEKIKIACTVSGPRPSKTFAFRSSAKLNCEFRLSPF 91
> CE20469
Length=240
Score = 33.9 bits (76), Expect = 0.068, Method: Composition-based stats.
Identities = 16/46 (34%), Positives = 27/46 (58%), Gaps = 1/46 (2%)
Query 17 SASGSAFVSLGNTKVYCSVFGPRPAGRSD-LQDRGFIKVDFRSSPF 61
+A GS ++ GNTKV C+V+GP S ++D+ I + ++ F
Sbjct 31 NAEGSCYLEHGNTKVLCAVYGPYEGKSSKRIEDKCAIVCQYSATKF 76
> Hs9506689
Length=245
Score = 33.9 bits (76), Expect = 0.076, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 24/50 (48%), Gaps = 3/50 (6%)
Query 15 LGSASGSAFVSLGNTKVYCSVFGP---RPAGRSDLQDRGFIKVDFRSSPF 61
A GSA++ GNTK V+GP R + L DR + + S+ F
Sbjct 32 FAQADGSAYIEQGNTKALAVVYGPHEIRGSRARALPDRALVNCQYSSATF 81
> At3g61620
Length=241
Score = 32.3 bits (72), Expect = 0.24, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 15 LGSASGSAFVSLGNTKVYCSVFGPR 39
+ A GSA +GNTKV +V+GPR
Sbjct 29 VSKADGSAVFEMGNTKVIAAVYGPR 53
> CE26236_1
Length=252
Score = 32.0 bits (71), Expect = 0.30, Method: Composition-based stats.
Identities = 15/56 (26%), Positives = 28/56 (50%), Gaps = 1/56 (1%)
Query 2 NKDIPPMHLQTLALGSASGSAFVSLGNTKVYCSVFGPRPAGRSDLQDRGFIKVDFR 57
N P+ ++ G+ GS + GNT+V + GP G+ + +DR I ++ +
Sbjct 34 NTAFRPLCVKCGVFGAQDGSGYAEFGNTRVLAQITGPDGDGKWE-EDRAKITIELK 88
> ECU10g0270
Length=188
Score = 30.8 bits (68), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 23/45 (51%), Gaps = 1/45 (2%)
Query 14 ALGSASGSAFVSLGNTKVYCSVFGPRPA-GRSDLQDRGFIKVDFR 57
+ +GS+ S NT V+C V GP A R + DR + V +R
Sbjct 10 VISHCTGSSRFSYNNTTVFCVVHGPSDAISRQEDPDRAILDVRWR 54
> CE26424
Length=434
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 24/48 (50%), Gaps = 0/48 (0%)
Query 13 LALGSASGSAFVSLGNTKVYCSVFGPRPAGRSDLQDRGFIKVDFRSSP 60
L +G+ G+A ++GNTKV +V S +G I +D SP
Sbjct 37 LVVGAEVGTAICTIGNTKVMAAVSAEIAEPSSMRPHKGVINIDVDLSP 84
> 7298022
Length=246
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 12/22 (54%), Positives = 16/22 (72%), Gaps = 0/22 (0%)
Query 20 GSAFVSLGNTKVYCSVFGPRPA 41
GSA++ GNTKV +V+GP A
Sbjct 38 GSAYMEQGNTKVLAAVYGPHQA 59
> 7303006
Length=281
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 1 NNKDIPPMHLQTLALGSASGSAFVSLGNTKVYCSV 35
+ +D PM L+T + +ASGSA + L NT + V
Sbjct 13 SRRDYRPMDLETGLVSNASGSARLRLANTDILVGV 47
> YPL167c
Length=1504
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 23/40 (57%), Gaps = 6/40 (15%)
Query 35 VFGPRPAGRSD----LQDRGFIKVDFRSSPFFQKNSAEVE 70
V+G P G D L+D GF K+D++ PFF N ++E
Sbjct 522 VYGEPPFGYQDILNKLEDEGFPKIDYK-DPFFS-NPVDLE 559
> At3g07750
Length=293
Score = 27.7 bits (60), Expect = 5.3, Method: Composition-based stats.
Identities = 22/57 (38%), Positives = 31/57 (54%), Gaps = 6/57 (10%)
Query 7 PMHLQTLALGSASGSAFVSLGNTKVYCSVFGP--RPAGRSDLQ-DRGFIKVDFRSSP 60
P++++T + A+GSA V +G T V SV RP S LQ D+G + V SP
Sbjct 31 PIYVETGVIPQANGSARVRIGGTDVIASVKAEIGRP---SSLQPDKGKVAVFIDCSP 84
> At5g14580
Length=991
Score = 27.3 bits (59), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 4 DIPPMHLQTLALGSASGSAFVSLGNTKVYCSVFGPRPA 41
++ P++ ++ L + GSA S G+T+V C+V PA
Sbjct 372 EVRPIYCESHYLPALHGSALFSRGDTQVLCTVTLGAPA 409
> YMR098c
Length=612
Score = 26.9 bits (58), Expect = 8.0, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 27/53 (50%), Gaps = 0/53 (0%)
Query 19 SGSAFVSLGNTKVYCSVFGPRPAGRSDLQDRGFIKVDFRSSPFFQKNSAEVEE 71
S SA LG+ V C+ + +S L+ F+K +F SS + + N E +E
Sbjct 132 SVSAVNKLGDFMVICTARSTKHCHKSFLELNKFLKHEFCSSAYVEGNFNERQE 184
Lambda K H
0.319 0.137 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1168763848
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40