bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0740_orf3
Length=288
Score E
Sequences producing significant alignments: (Bits) Value
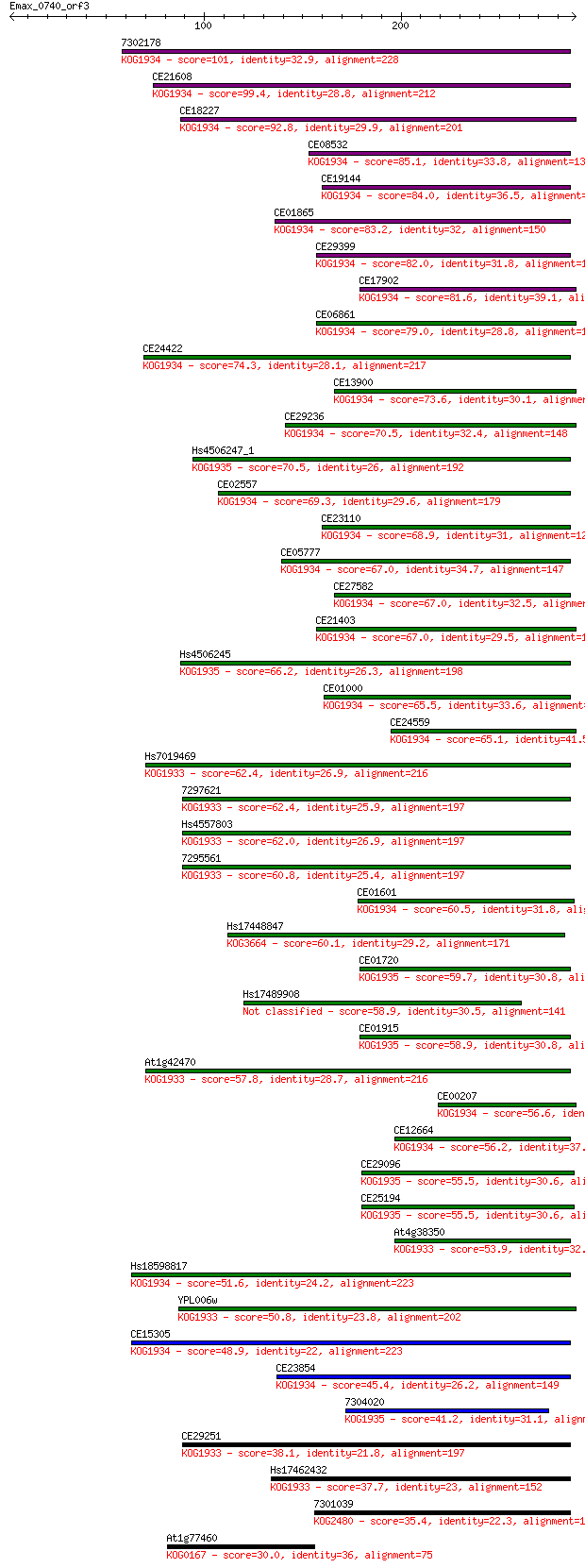
7302178 101 2e-21
CE21608 99.4 9e-21
CE18227 92.8 8e-19
CE08532 85.1 1e-16
CE19144 84.0 4e-16
CE01865 83.2 7e-16
CE29399 82.0 1e-15
CE17902 81.6 2e-15
CE06861 79.0 1e-14
CE24422 74.3 3e-13
CE13900 73.6 5e-13
CE29236 70.5 4e-12
Hs4506247_1 70.5 4e-12
CE02557 69.3 8e-12
CE23110 68.9 1e-11
CE05777 67.0 4e-11
CE27582 67.0 5e-11
CE21403 67.0 5e-11
Hs4506245 66.2 8e-11
CE01000 65.5 1e-10
CE24559 65.1 2e-10
Hs7019469 62.4 1e-09
7297621 62.4 1e-09
Hs4557803 62.0 1e-09
7295561 60.8 3e-09
CE01601 60.5 4e-09
Hs17448847 60.1 6e-09
CE01720 59.7 7e-09
Hs17489908 58.9 1e-08
CE01915 58.9 1e-08
At1g42470 57.8 2e-08
CE00207 56.6 6e-08
CE12664 56.2 9e-08
CE29096 55.5 1e-07
CE25194 55.5 1e-07
At4g38350 53.9 3e-07
Hs18598817 51.6 2e-06
YPL006w 50.8 3e-06
CE15305 48.9 1e-05
CE23854 45.4 2e-04
7304020 41.2 0.003
CE29251 38.1 0.024
Hs17462432 37.7 0.031
7301039 35.4 0.15
At1g77460 30.0 6.0
> 7302178
Length=1061
Score = 101 bits (251), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 75/235 (31%), Positives = 116/235 (49%), Gaps = 19/235 (8%)
Query 58 EDAFVLG--SIECITSASALMLHYDADGRPMHEERNATWERLLIQVLKANQTLGSLR--- 112
ED +V+ +I+ + +A DA G A WE ++V+ + G +
Sbjct 91 EDNYVISVPAIQLVYFVTADTKRQDAKG--------AEWEETFLRVVGNAENSGQFKHIS 142
Query 113 -AYFQAFRSRDDELKASTAESKDVV-YVVFTFFLLASYSTALNFSCDLYRSKLFSSLMGF 170
+YF A R+ D EL+ +T K VV Y TF L+ +S D RSK F LMG
Sbjct 143 VSYF-ASRTLDHELEKNT---KTVVPYFSSTFLLMGLFSIITCMMGDAVRSKPFLGLMGN 198
Query 171 FAAFMGLGAGMGIVCYMKVAMVPTVLICPFLVLGIGVDDVFVLLNAYCMGYTTHDPEQRC 230
+A M A G+ Y + + L PFL++GIG+DD FV+L + +R
Sbjct 199 VSAIMATLAAFGLAMYCGIEFIGINLAAPFLMIGIGIDDTFVMLAGWRRTKAKMPVAERM 258
Query 231 IDTLKTSGLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMALLLGYIFVLTFF 285
+ + + ITIT++T+ ISF +G S + S+R FC YS A+ +++ +TFF
Sbjct 259 GLMMSEAAVSITITSVTDFISFLIGIISPFRSVRIFCTYSVFAVCFTFLWHITFF 313
> CE21608
Length=800
Score = 99.4 bits (246), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 61/212 (28%), Positives = 108/212 (50%), Gaps = 3/212 (1%)
Query 74 ALMLHYDADGRPMHEERNATWERLLIQVLKANQTLGSLRAYFQAFRSRDDELKASTAESK 133
A+ H + M+E WE+ L + + +R Y + +E++ + +
Sbjct 111 AINFHAIYNNETMYEIMKE-WEQKLFAYTLSTENDPLIRVYVTSEGLVSEEVRRTGILAM 169
Query 134 DVVYVVFTFFLLASYSTALNFSCDLYRSKLFSSLMGFFAAFMGLGAGMGIVCYMKVAMVP 193
++ V TF ++ +++ D +SK F + +G + L A G + +M +P
Sbjct 170 PLMGV--TFLIILAFTILTTLKRDPVKSKPFEAFLGVICPILSLCASFGHLFWMGFEYLP 227
Query 194 TVLICPFLVLGIGVDDVFVLLNAYCMGYTTHDPEQRCIDTLKTSGLGITITTLTNLISFG 253
V + PFL+L IGVDDVF+ ++A+ H R +TL +G I+IT+LTNL+SF
Sbjct 228 IVTVVPFLILSIGVDDVFIFIHAWHRTPYKHSVRDRMAETLADAGPSISITSLTNLLSFA 287
Query 254 VGSFSTYLSIRNFCVYSAMALLLGYIFVLTFF 285
+G F+ +I FCV+ + A++ YI+ + FF
Sbjct 288 IGIFTPTPAIYTFCVFISTAVIYDYIYQIFFF 319
> CE18227
Length=840
Score = 92.8 bits (229), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 60/201 (29%), Positives = 110/201 (54%), Gaps = 5/201 (2%)
Query 88 EERNATWERLLIQVLKANQTLGSLRAYFQAFRSRDDELKASTAESKDVVYVVFTFFLLAS 147
EE + +E+ L +L+ T + + F +D+ K ST + ++ T LL
Sbjct 221 EEMSKIFEQSLTALLENQTTFDT--SMFSLSILKDEMQKNSTYT---MPFISLTILLLLC 275
Query 148 YSTALNFSCDLYRSKLFSSLMGFFAAFMGLGAGMGIVCYMKVAMVPTVLICPFLVLGIGV 207
++ A + + SK +++G + M + + G++ + V + V + PF+ L IGV
Sbjct 276 FTVASCMTDNWVTSKPIEAMIGILVSSMAIVSAGGLLFALGVPFINQVTVMPFIALAIGV 335
Query 208 DDVFVLLNAYCMGYTTHDPEQRCIDTLKTSGLGITITTLTNLISFGVGSFSTYLSIRNFC 267
DDV+V+L A+ T D ++R L+ +G IT+T+LT+++SFG+G++ST +I FC
Sbjct 336 DDVYVMLGAWQDTRRTLDAKKRMGLALEEAGSAITVTSLTSVLSFGIGTYSTTPAIAIFC 395
Query 268 VYSAMALLLGYIFVLTFFFPV 288
+ A+A++ + + LTFF V
Sbjct 396 KFIALAIMFDWFYQLTFFAAV 416
> CE08532
Length=933
Score = 85.1 bits (209), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 45/133 (33%), Positives = 80/133 (60%), Gaps = 0/133 (0%)
Query 153 NFSCDLYRSKLFSSLMGFFAAFMGLGAGMGIVCYMKVAMVPTVLICPFLVLGIGVDDVFV 212
+FS D SK S++G +A + + G+G + M + V + PFLVL +GVD++F+
Sbjct 290 SFSIDWVLSKPILSILGVVSAGIAILTGVGFLSLMGMPYNDIVGVMPFLVLAVGVDNMFL 349
Query 213 LLNAYCMGYTTHDPEQRCIDTLKTSGLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAM 272
++ A TH +R + L + + I IT+ T+++SFGVG+ +T +++ FCVY+ +
Sbjct 350 MVAAVRRTSRTHTVHERMGECLADAAVSILITSSTDVLSFGVGAITTIPAVQIFCVYTGV 409
Query 273 ALLLGYIFVLTFF 285
A+ +I+ +TFF
Sbjct 410 AIFFAFIYQITFF 422
> CE19144
Length=1003
Score = 84.0 bits (206), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 46/126 (36%), Positives = 76/126 (60%), Gaps = 0/126 (0%)
Query 160 RSKLFSSLMGFFAAFMGLGAGMGIVCYMKVAMVPTVLICPFLVLGIGVDDVFVLLNAYCM 219
RS+ + ++ G +A M + + +GI+ M PF+V +GVD+VF+LL+A+
Sbjct 319 RSRPWLAIGGVISAAMAIASAVGILLLAGYGMTSVAYSMPFIVFSVGVDNVFILLSAWRS 378
Query 220 GYTTHDPEQRCIDTLKTSGLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMALLLGYI 279
+T E R +T + + IT+T+LT+LISFGVG + + S++ FC Y+ A++ YI
Sbjct 379 TSSTETLEHRMKETFADAAVSITVTSLTDLISFGVGCATPFPSVQMFCAYAVAAVIFTYI 438
Query 280 FVLTFF 285
+ LTFF
Sbjct 439 YQLTFF 444
> CE01865
Length=1015
Score = 83.2 bits (204), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 48/160 (30%), Positives = 89/160 (55%), Gaps = 10/160 (6%)
Query 136 VYVVFTFFLLASYSTALNFSCDLY----RSKLFSSLMGFFAAFMGLGAGMGIVCYMKVAM 191
V V+FT F + ++ S L+ RSK +L G ++ + + +G+G++ + +
Sbjct 341 VLVLFTAFYSVKWYFRMDHSWPLHIDWLRSKPMLALGGVLSSVLAILSGIGLLLWFGMFF 400
Query 192 VPTVLICPFLVLGIGVDDVFVLLNAYC---MGYTTHDPE---QRCIDTLKTSGLGITITT 245
LI PFLVL IGVDD+F+ + A+ M Y P+ +R I+ + S + I IT+
Sbjct 401 AEITLIAPFLVLSIGVDDMFIAVAAWHNTEMKYPGRSPKVMKKRMIEAMSESAVAIFITS 460
Query 246 LTNLISFGVGSFSTYLSIRNFCVYSAMALLLGYIFVLTFF 285
T+++SFG G+ + ++++ FC +A + +++ +TFF
Sbjct 461 FTDVLSFGAGTITDIIAVQGFCAMTAACMFFTFLYQITFF 500
> CE29399
Length=870
Score = 82.0 bits (201), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 41/129 (31%), Positives = 77/129 (59%), Gaps = 0/129 (0%)
Query 157 DLYRSKLFSSLMGFFAAFMGLGAGMGIVCYMKVAMVPTVLICPFLVLGIGVDDVFVLLNA 216
D +K S++G A MG+ + MG++ Y+++ + + PFLV+ +G D++F+++ +
Sbjct 267 DWVVTKPILSVLGVSNAGMGIASAMGMLTYLEIQYNDIIAVMPFLVVAVGTDNMFLMVAS 326
Query 217 YCMGYTTHDPEQRCIDTLKTSGLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMALLL 276
+QR + + + + I IT LT+ +SFGVG+ +T +++ FC+Y+ ALLL
Sbjct 327 LKRTDRNLKYDQRIAECMADAAVSILITALTDALSFGVGTITTIPAVQIFCIYTMCALLL 386
Query 277 GYIFVLTFF 285
+ + LTFF
Sbjct 387 TFAYQLTFF 395
> CE17902
Length=820
Score = 81.6 bits (200), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 43/110 (39%), Positives = 62/110 (56%), Gaps = 0/110 (0%)
Query 179 AGMGIVCYMKVAMVPTVLICPFLVLGIGVDDVFVLLNAYCMGYTTHDPEQRCIDTLKTSG 238
A G +C+M + PFLVLGIGVDD F+LL+ + D +R + G
Sbjct 285 ASFGAICWMGFPSFSIQCVTPFLVLGIGVDDAFILLHRWKHHIAITDTPRRLEQVIVDVG 344
Query 239 LGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMALLLGYIFVLTFFFPV 288
ITIT+LTN+I+FG+G + + FC+ +++ALLL YIF T P+
Sbjct 345 PSITITSLTNIIAFGIGFLTPTPQMSLFCLTASLALLLDYIFTYTILAPI 394
> CE06861
Length=956
Score = 79.0 bits (193), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 38/132 (28%), Positives = 68/132 (51%), Gaps = 0/132 (0%)
Query 157 DLYRSKLFSSLMGFFAAFMGLGAGMGIVCYMKVAMVPTVLICPFLVLGIGVDDVFVLLNA 216
D RSK + G M G++ + V + PFL+L IG+DD+F++
Sbjct 319 DWVRSKPLVAAAGLMTPIMATVTSFGLILWCGFLYNAIVNVSPFLILCIGIDDLFIMCAE 378
Query 217 YCMGYTTHDPEQRCIDTLKTSGLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMALLL 276
+ H PE+R TL + + I+IT+LT++ +F +G ++T ++ FC+Y+ +
Sbjct 379 WHRTNPQHSPEKRIGKTLSEAAVAISITSLTDIATFAMGCYTTLPGVQMFCMYTCVQCFF 438
Query 277 GYIFVLTFFFPV 288
Y++ + F PV
Sbjct 439 CYVYQIIFLGPV 450
> CE24422
Length=860
Score = 74.3 bits (181), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 61/222 (27%), Positives = 106/222 (47%), Gaps = 10/222 (4%)
Query 69 ITSASALMLHYDADGR-PMHEERNATWERLLIQVLKANQTLGSLRAYFQAFRSRD-DELK 126
+T A ++LHY D P +E + + L Y R+R+ +E +
Sbjct 182 VTEAKMVVLHYMVDTSIPNGRALATDFENKVRAFFASISDLSHHIKYSVLSRTRELEEQR 241
Query 127 ASTAESK---DVVYVVFTFFLLASYSTALNFSCDLYRSKLFSSLMGFFAAFMGLGAGMGI 183
A T S + VV T F+L T + F LY S+ + S++G + M L G+
Sbjct 242 AITITSLPFLGLTIVVLTCFMLV---TLVRFP--LYTSQHWQSIVGVLSPGMALWTTTGL 296
Query 184 VCYMKVAMVPTVLICPFLVLGIGVDDVFVLLNAYCMGYTTHDPEQRCIDTLKTSGLGITI 243
+ + + + PFLV+ IG+DD F++L + E R ++ SG +T+
Sbjct 297 LWGIGYPFSNILTVVPFLVVTIGIDDAFLILAGWRQSTKGESLEVRLGQSVAISGASVTV 356
Query 244 TTLTNLISFGVGSFSTYLSIRNFCVYSAMALLLGYIFVLTFF 285
T++T++ F G FS ++ FC+Y+ +AL + +I+ +TFF
Sbjct 357 TSVTDVACFATGLFSNMPVVQLFCLYTTVALAIDFIYQMTFF 398
> CE13900
Length=881
Score = 73.6 bits (179), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 37/123 (30%), Positives = 68/123 (55%), Gaps = 0/123 (0%)
Query 166 SLMGFFAAFMGLGAGMGIVCYMKVAMVPTVLICPFLVLGIGVDDVFVLLNAYCMGYTTHD 225
+++G M G G + L+ PFL++G+G DDVF++++A +
Sbjct 216 AILGIAGPLMATGTAFGFLFLFGFPFNSITLVMPFLIIGVGCDDVFIIIHAMRKTDKSES 275
Query 226 PEQRCIDTLKTSGLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMALLLGYIFVLTFF 285
E R +T++ +G IT+T+ TN +SF +G + +I FC+Y+ +A+ + +++ LTFF
Sbjct 276 LEDRIAETMEEAGPSITVTSATNCLSFAIGIATPTPAISLFCLYTCVAVAVDFVYQLTFF 335
Query 286 FPV 288
V
Sbjct 336 VAV 338
> CE29236
Length=968
Score = 70.5 bits (171), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 48/150 (32%), Positives = 77/150 (51%), Gaps = 3/150 (2%)
Query 141 TFFLLASYSTALNFSCDLYRSKLFSSLMGFFAAFMGLGAGMGIVCYMKVAMVPTVLICPF 200
TF L TA+ +C L + KL SL + + + GIV + + +L+ PF
Sbjct 336 TFVSLCVILTAVYHNC-LDQGKLLISLGAVLCPILAITSTYGIVSILGMRTNSFMLVMPF 394
Query 201 LVLGIGVDDVFVLLNAYCMGYTTH--DPEQRCIDTLKTSGLGITITTLTNLISFGVGSFS 258
LV+GIGVD F++++++ R ++ G ITIT+LT+ +SF +G+ +
Sbjct 395 LVMGIGVDSCFLMIHSWQKERRAQASGTGNRLGMVYESVGPSITITSLTDFLSFAIGALA 454
Query 259 TYLSIRNFCVYSAMALLLGYIFVLTFFFPV 288
R FC+ S++AL L YI L F P+
Sbjct 455 PTPETRLFCIASSIALALTYILQLVLFGPI 484
> Hs4506247_1
Length=1390
Score = 70.5 bits (171), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 50/196 (25%), Positives = 97/196 (49%), Gaps = 6/196 (3%)
Query 94 WERLLIQVLKANQTLGSLRAYFQ-AFRSRDDELKASTAESKDVVYVVFTFFLLASYSTAL 152
W+R ++V+ + S + + DD LK+ + S V+ V + L+ +Y+
Sbjct 399 WQRTYVEVVHQSVAQNSTQKVLSFTTTTLDDILKSFSDVS--VIRVASGYLLMLAYACLT 456
Query 153 NFSCDLYRSKLFSSLMGFFAAFMGLGAGMGIVCYMKVAM-VPTVLICPFLVLGIGVDDVF 211
D +S+ L G + + AG+G+ + ++ T + PFL LG+GVDDVF
Sbjct 457 MLRWDCSKSQGAVGLAGVLLVALSVAAGLGLCSLIGISFNAATTQVLPFLALGVGVDDVF 516
Query 212 VLLNAYC-MGYTTHDP-EQRCIDTLKTSGLGITITTLTNLISFGVGSFSTYLSIRNFCVY 269
+L +A+ G P E R + LK +G + +T+++N+ +F + + ++R F +
Sbjct 517 LLAHAFSETGQNKRIPFEDRTGECLKRTGASVALTSISNVTAFFMAALIPIPALRAFSLQ 576
Query 270 SAMALLLGYIFVLTFF 285
+A+ ++ + VL F
Sbjct 577 AAVVVVFNFAMVLLIF 592
> CE02557
Length=1037
Score = 69.3 bits (168), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 53/202 (26%), Positives = 90/202 (44%), Gaps = 24/202 (11%)
Query 107 TLGSLRAYFQAFRSRDDELKASTAESKDVVY-------------------VVFTF---FL 144
T + RAY+ A+R + +E S +E+ D+ + V F F L
Sbjct 371 TAENNRAYY-AYRQKLNEYWESISENIDLQFIPHNDKAMDDEMLEIIKTTVPFAFPATIL 429
Query 145 LASYSTALNFSCDLYRSKLFSSLMGFFAAFMGLGAGMGIVCYMKVAMVPTVLICPFLVLG 204
L + + N+S D +SK +G + L G+ + P PFLVL
Sbjct 430 LTIFVLSANYSSDRTKSKPIEMCVGVWCVIFALIITFGVFFFFGAKFNPVTSTMPFLVLA 489
Query 205 IGVDDVFVLLNAYCMGYTTHDPEQRCIDTLKTSGLGITITTLTNLISFGVGSF-STYLSI 263
+GVDD F+++ A+ H P R + +G IT+T+ TN F +G F + ++
Sbjct 490 VGVDDDFLMMAAWRECDRKHSPAIRLGFVMADAGASITVTSFTNFFCFFLGWFMCSTPAV 549
Query 264 RNFCVYSAMALLLGYIFVLTFF 285
+FC+ +A + Y+ +TF+
Sbjct 550 ADFCIITAAGVFFDYLMQITFY 571
> CE23110
Length=859
Score = 68.9 bits (167), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 39/126 (30%), Positives = 70/126 (55%), Gaps = 0/126 (0%)
Query 160 RSKLFSSLMGFFAAFMGLGAGMGIVCYMKVAMVPTVLICPFLVLGIGVDDVFVLLNAYCM 219
SK + +G + +G+V +A ++ FLV+ IG+DD+F++L A+
Sbjct 247 ESKPLEACLGALIPVLSGITTIGMVSATGLAFQSIIVSTLFLVIAIGIDDIFIILAAWHR 306
Query 220 GYTTHDPEQRCIDTLKTSGLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMALLLGYI 279
+ +R T++ +G +T+TT+TNL+SFG G ST ++ F +YS++A ++ YI
Sbjct 307 TDKHLEIPERMALTVQEAGCSMTVTTVTNLVSFGNGVLSTTPVLQTFAIYSSVASVVCYI 366
Query 280 FVLTFF 285
+ L F
Sbjct 367 YQLVIF 372
> CE05777
Length=955
Score = 67.0 bits (162), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 51/173 (29%), Positives = 86/173 (49%), Gaps = 29/173 (16%)
Query 139 VFTFFLLASYSTALNFS--------CDLYRSKLFSSLMGFFAAFMGLGAGMGIVCYMKVA 190
V +FF+LA Y+ +F+ D SK + + G F+ + + + G + + V
Sbjct 270 VVSFFVLAMYALVSSFTLKSSSATKIDWISSKPWLAAAGMFSTVLSIISAFGFLFILGVR 329
Query 191 MVPTVLICPFLVLG---------------IGVDDVFVLLNAYCMGYTTHD---PEQRCID 232
I PFL++G IG+DD+F L+NA C T+ PE R
Sbjct 330 YNVINTIIPFLIIGEFQSFKNMEKNQIPAIGIDDMF-LMNA-CWDQTSKSLSVPE-RMSK 386
Query 233 TLKTSGLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMALLLGYIFVLTFF 285
TL +G+ +TIT +T+++SF +G + I+ FC+Y+ +++ Y + LTFF
Sbjct 387 TLSHAGVAVTITNVTDVMSFAIGCITDLPGIQFFCIYACVSVAFSYFYQLTFF 439
> CE27582
Length=914
Score = 67.0 bits (162), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 39/132 (29%), Positives = 71/132 (53%), Gaps = 13/132 (9%)
Query 166 SLMGFFAAFMGLGAGMGIVCYMKVAMVPTVLICPFLVLGIGVDDVFVLLNAYCMGYTTH- 224
++M FM GA +G + ++ + + PFLVL IGVDD +++ NA+ T H
Sbjct 311 AVMACVCPFMACGASLGAMFFIGFRFGSILCVTPFLVLAIGVDDSYLMANAW-QRITCHR 369
Query 225 -----------DPEQRCIDTLKTSGLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMA 273
+ + R + +G ITITT+TN ++FG+G+ + I+ F + +A+A
Sbjct 370 RKHARFESVNVELKNRITEMFIETGPSITITTITNALAFGIGATTPAAEIQLFSIGNALA 429
Query 274 LLLGYIFVLTFF 285
++ ++F +TF+
Sbjct 430 VITDFVFTITFY 441
> CE21403
Length=936
Score = 67.0 bits (162), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 39/132 (29%), Positives = 68/132 (51%), Gaps = 0/132 (0%)
Query 157 DLYRSKLFSSLMGFFAAFMGLGAGMGIVCYMKVAMVPTVLICPFLVLGIGVDDVFVLLNA 216
D SK + G + + G++ V V + PFL L IG+DD F++L A
Sbjct 326 DWVLSKPLLGICGVLVTMCAIISSTGLLMLFNVTFVDMCTVMPFLSLTIGIDDTFLMLAA 385
Query 217 YCMGYTTHDPEQRCIDTLKTSGLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMALLL 276
+ E+R ++ + + I+IT+LT+ ++F +GS + ++ FC YS+ A+L
Sbjct 386 WHETDRNLPYEKRIEKAMRHAAVSISITSLTDALAFLIGSIAPLPAVIYFCYYSSAAILF 445
Query 277 GYIFVLTFFFPV 288
+++VLT F V
Sbjct 446 IFLYVLTMFVAV 457
> Hs4506245
Length=1203
Score = 66.2 bits (160), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 52/206 (25%), Positives = 99/206 (48%), Gaps = 12/206 (5%)
Query 88 EERNAT----WERLLIQVLKANQTLG-SLRAYFQAFRSR--DDELKASTAESKDVVYVVF 140
EE+ +T W+R +Q+ A + L + AF S DD L A + S VV
Sbjct 345 EEQASTVLQAWQRRFVQL--AQEALPENASQQIHAFSSTTLDDILHAFSEVS--AARVVG 400
Query 141 TFFLLASYSTALNFSCDLYRSKLFSSLMGFFAAFMGLGAGMGIVCYMKVAM-VPTVLICP 199
+ L+ +Y+ D +S+ L G + + +G+G+ + + T + P
Sbjct 401 GYLLMLAYACVTMLRWDCAQSQGSVGLAGVLLVALAVASGLGLCALLGITFNAATTQVLP 460
Query 200 FLVLGIGVDDVFVLLNAYCMGYTTHDPEQRCIDTLKTSGLGITITTLTNLISFGVGSFST 259
FL LGIGVDDVF+L +A+ ++R + L+ +G + +T++ N+ +F + +
Sbjct 461 FLALGIGVDDVFLLAHAFTEALPGTPLQERMGECLQRTGTSVVLTSINNMAAFLMAALVP 520
Query 260 YLSIRNFCVYSAMALLLGYIFVLTFF 285
++R F + +A+ + ++ V+ F
Sbjct 521 IPALRAFSLQAAIVVGCTFVAVMLVF 546
> CE01000
Length=890
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 42/132 (31%), Positives = 70/132 (53%), Gaps = 7/132 (5%)
Query 161 SKLFSSLMGFFAAFMGLGAGMGIVCYMKVAMVPTVLICPFLVLGIGVDDVFVLLNAYCM- 219
+K+ S++ FM G MGI+ + V P + I PFLVL I VDD F++L+A+
Sbjct 296 NKVALSIIACINPFMACGTAMGILFFCGVTFSPIMCITPFLVLAISVDDSFLMLHAWNRL 355
Query 220 -GYTT---HDP--EQRCIDTLKTSGLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMA 273
+ T P E + L +G I+I+ TN+++F +G+ ++ IR FC +A A
Sbjct 356 ESWRTAPLDKPMREHMMGEVLVETGPAISISAFTNMLAFTIGAITSPPEIRIFCFGNAAA 415
Query 274 LLLGYIFVLTFF 285
+ + + TF+
Sbjct 416 IFMDMFYQATFY 427
> CE24559
Length=909
Score = 65.1 bits (157), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 39/98 (39%), Positives = 57/98 (58%), Gaps = 8/98 (8%)
Query 195 VLICPFLVLGIGVDDVFVLLNAYCMGYTTHDPE----QRCIDTLKTSGLGITITTLTNLI 250
++I PFL+ GIGV+D F+ L + H P+ +R L +G IT TTLTN+I
Sbjct 356 MMIMPFLINGIGVNDAFLTLQ----NWLQHSPDLPSGKRLGYMLAEAGPSITTTTLTNVI 411
Query 251 SFGVGSFSTYLSIRNFCVYSAMALLLGYIFVLTFFFPV 288
F +G + + FC+ A++LLL Y++ LTFF PV
Sbjct 412 VFLIGWMNPTEEMSIFCLGCAISLLLAYVYTLTFFCPV 449
> Hs7019469
Length=1359
Score = 62.4 bits (150), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 58/230 (25%), Positives = 109/230 (47%), Gaps = 17/230 (7%)
Query 70 TSASALMLHYDADGRPMHEERNAT---WERLLIQVLKANQT--LGSLRAYFQAFRSRDDE 124
+ A AL++ + + P + R A WE ++ ++A Q G + F A RS +DE
Sbjct 565 SEAEALIMTFSLNNYPAGDPRLAQAKLWEEAFLEEMRAFQRRMAGMFQVTFMAERSLEDE 624
Query 125 LKASTAESKDV---VYVVFTFFL---LASYSTALNFSCDLYRSKLFSSLMGFFAAFMGLG 178
+ +TAE + Y+V ++ L SYS+ +S + SK L G +
Sbjct 625 INRTTAEDLPIFATSYIVIFLYISLALGSYSS---WSRVMVDSKATLGLGGVAVVLGAVM 681
Query 179 AGMGIVCYMKVAMVPTVL-ICPFLVLGIGVDDVFVLLNAYC-MGYTTHDPEQRCID-TLK 235
A MG Y+ + +L + PFLVL +G D++F+ + Y + +P + I L
Sbjct 682 AAMGFFSYLGIRSSLVILQVVPFLVLSVGADNIFIFVLEYQRLPRRPGEPREVHIGRALG 741
Query 236 TSGLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMALLLGYIFVLTFF 285
+ + +L+ I F +G+ + ++R F + S +A++L ++ ++ F
Sbjct 742 RVAPSMLLCSLSEAICFFLGALTPMPAVRTFALTSGLAVILDFLLQMSAF 791
> 7297621
Length=1157
Score = 62.4 bits (150), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 51/209 (24%), Positives = 103/209 (49%), Gaps = 15/209 (7%)
Query 89 ERNATWERLLIQVLKANQTLGSLRAY----FQAFRSRDDELKASTAESKDVVYVVFTFFL 144
E TWE+ ++ + N T ++ Y F + RS +DEL + DV+ ++ ++ +
Sbjct 446 ENALTWEKKFVEFM-TNYTKNNMSQYMDIAFTSERSIEDELNRES--QSDVLTILVSYLI 502
Query 145 LASY-STALNFSCDLYR----SKLFSSLMGFFAAFMGLGAGMGIVCYMKV-AMVPTVLIC 198
+ Y + +L + R SK+ + G + + +G+ Y+ + A + V +
Sbjct 503 MFMYIAISLGHVKEFKRVFIDSKITLGIGGVIIVLASVVSSVGVFGYIGLPATLIIVEVI 562
Query 199 PFLVLGIGVDDVFVLLNAYCMGYTTHDP--EQRCIDTLKTSGLGITITTLTNLISFGVGS 256
PFLVL +GVD++F+L+ + + EQ+ L G + +T+L+ F +G
Sbjct 563 PFLVLAVGVDNIFILVQTHQRDQRKPNETLEQQVGRILGKVGPSMLLTSLSESFCFFLGG 622
Query 257 FSTYLSIRNFCVYSAMALLLGYIFVLTFF 285
S ++R F +Y+ +AL++ ++ +T F
Sbjct 623 LSDMPAVRAFALYAGVALIIDFLLQITCF 651
> Hs4557803
Length=1278
Score = 62.0 bits (149), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 53/208 (25%), Positives = 99/208 (47%), Gaps = 17/208 (8%)
Query 89 ERNATWERLLIQVLKANQTLGSLRAYFQAFRSRDDELKASTAESKDVVYVVFTF---FLL 145
+R WE+ I +K N +L F A RS +DEL + DV VV ++ FL
Sbjct 578 QRAQAWEKEFINFVK-NYKNPNLTISFTAERSIEDELNRES--DSDVFTVVISYAIMFLY 634
Query 146 ASYSTALNFSCD--LYRSKLFSSLMGFFAAFMGLGAGMGIVCYMKVAMVPTVL-ICPFLV 202
S + SC L SK+ + G + +G+ Y+ + + V+ + PFLV
Sbjct 635 ISLALGHIKSCRRLLVDSKVSLGIAGILIVLSSVACSLGVFSYIGLPLTLIVIEVIPFLV 694
Query 203 LGIGVDDVFVLLNAY-----CMGYTTHDPEQRCIDTLKTSGLGITITTLTNLISFGVGSF 257
L +GVD++F+L+ AY G T R + + S + +++ + ++F +G+
Sbjct 695 LAVGVDNIFILVQAYQRDERLQGETLDQQLGRVLGEVAPS---MFLSSFSETVAFFLGAL 751
Query 258 STYLSIRNFCVYSAMALLLGYIFVLTFF 285
S ++ F +++ +A+ + ++ +T F
Sbjct 752 SVMPAVHTFSLFAGLAVFIDFLLQITCF 779
> 7295561
Length=1223
Score = 60.8 bits (146), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 50/205 (24%), Positives = 93/205 (45%), Gaps = 11/205 (5%)
Query 89 ERNATWERLLIQVLKANQTLGSLRAYFQAFRSRDDELKASTAESKDVVYVVFTFFLLASY 148
E N WE+L + L+ ++ AY +D ++ S E VV F+ +
Sbjct 519 EPNMKWEKLFVDFLRDYKSDRLDIAYMAERSIQDAIVELSEGEVSTVVISYVVMFVYVAI 578
Query 149 STALNFSCD--LYRSKLFSSLMGFFAAFMGLGAGMGIVCYMKVAM-VPTVLICPFLVLGI 205
+ SC L S++ ++ G + +G Y+ V + + + PFLVL +
Sbjct 579 ALGHIRSCRGFLRESRIMLAIGGIVIVLASVVCSLGFWGYLDVTTTMLAIEVIPFLVLAV 638
Query 206 GVDDVFVLLNAY-----CMGYTTHDPEQRCIDTLKTSGLGITITTLTNLISFGVGSFSTY 260
GVD++F++++ Y TTH+ I + G I T + + F +G S
Sbjct 639 GVDNIFIMVHTYQRLDHSKFKTTHEAIGEAIGQV---GPSILQTAGSEMACFAIGCISDM 695
Query 261 LSIRNFCVYSAMALLLGYIFVLTFF 285
+++ F +Y+A+A+LL ++ +T F
Sbjct 696 PAVKTFAMYAAIAILLDFLLQITAF 720
> CE01601
Length=690
Score = 60.5 bits (145), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 35/114 (30%), Positives = 60/114 (52%), Gaps = 4/114 (3%)
Query 178 GAGMGIVCYMKVAMVPTVLICPFLVLGIGVDDVFVLLNAY----CMGYTTHDPEQRCIDT 233
GA I+ ++ + I PFL+LGIGVDD F+LLN + + E +
Sbjct 145 GACFAILGWVGHPFNSIMCITPFLILGIGVDDAFLLLNCWRREEGKDKSAKQAENQLARV 204
Query 234 LKTSGLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMALLLGYIFVLTFFFP 287
++ + IT+LTN ++FGVG + + +FC+ +A+A++L ++ F P
Sbjct 205 IREISPSMAITSLTNTMAFGVGFLAPTPQMSSFCLGTALAIVLDFLLEFLIFVP 258
> Hs17448847
Length=1392
Score = 60.1 bits (144), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 50/191 (26%), Positives = 89/191 (46%), Gaps = 23/191 (12%)
Query 112 RAYFQAFRSRDDELKASTAESKDVVYVVFTFFLLASYSTALNFSCDLYRSKLFSSLMG-- 169
RA FQ+F + A + SK V V++ L Y F+ D+ + + SS +
Sbjct 419 RAKFQSFVVTYVAMLAKQSTSK--VQVLYGGTDLFDYEVRRTFNNDMLLAFISSSCIAAL 476
Query 170 --------FFAAFMGLGAGMGIVCYMKVAMVPTVL----------ICPFLVLGIGVDDVF 211
F +F G+ A +G+ C + + + V + F+++GIGVDDVF
Sbjct 477 VYILTSCSVFLSFFGI-ASIGLSCLVALFLYHVVFGIQYLGILNGVAAFVIVGIGVDDVF 535
Query 212 VLLNAYCMGYTTHDPEQRCIDTLKTSGLGITITTLTNLISFGVGSFSTYLSIRNFCVYSA 271
V +N Y DP+ R I T++T+G T+LT ++ FS ++ +F ++ +
Sbjct 536 VFINTYRQATHLEDPQLRMIHTVQTAGKATFFTSLTTAAAYAANVFSQIPAVHDFGLFMS 595
Query 272 MALLLGYIFVL 282
+ + ++ VL
Sbjct 596 LIVSCCWLAVL 606
> CE01720
Length=1405
Score = 59.7 bits (143), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 33/108 (30%), Positives = 58/108 (53%), Gaps = 1/108 (0%)
Query 179 AGMGIVCYMKVAM-VPTVLICPFLVLGIGVDDVFVLLNAYCMGYTTHDPEQRCIDTLKTS 237
AG+G+ + + T I PFL LGIGVD++F+LL+ Y ++ +
Sbjct 700 AGLGLATWFGIEFNAATTQIVPFLTLGIGVDNMFMLLHNYRDVVKLAGGHAEMAILMRET 759
Query 238 GLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMALLLGYIFVLTFF 285
G+ I T++ N++SF G+ ++R+FC S++ L +I +LT +
Sbjct 760 GMSILCTSINNILSFLTGTLLPIPALRSFCAQSSILLTFNFIAILTIY 807
> Hs17489908
Length=674
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 43/144 (29%), Positives = 80/144 (55%), Gaps = 5/144 (3%)
Query 120 SRDDELKASTAESKDVVYVVFTFFLLASYSTALNFSCDLYRSKLFSSLMGFFAAFMGLGA 179
SR E +A++ V ++ + +L + ++ F C R+K+ + G +AF+ + +
Sbjct 384 SRQLEFEATSVTVIPVFHLAYILIILFAVTSCFRFDC--IRNKMCVAAFGVISAFLAVVS 441
Query 180 GMGIVCYMKVAMVPTVLICPFLVLGIGVDDVFVLLNAYCMGYTTHDPEQRCIDTLKTSGL 239
G G++ ++ V V V PFL+LG+GVDD+F++++A+ D +R + + +
Sbjct 442 GFGLLLHIGVPFVIIVANSPFLILGVGVDDMFIMISAWHKTNLADDIRERMSNVYSKAAV 501
Query 240 GITITTLTNLISFGVG---SFSTY 260
ITITT+TN+++ G SFS Y
Sbjct 502 SITITTITNILALYTGIMSSFSIY 525
> CE01915
Length=714
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 33/108 (30%), Positives = 58/108 (53%), Gaps = 1/108 (0%)
Query 179 AGMGIVCYMKVAM-VPTVLICPFLVLGIGVDDVFVLLNAYCMGYTTHDPEQRCIDTLKTS 237
AG+G+ + + T I PFL LGIGVD++F+LL+ Y ++ +
Sbjct 203 AGIGLATWFGIEFNAATTQIVPFLTLGIGVDNMFMLLHNYRDVVKLAGGHAEMAILMRET 262
Query 238 GLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMALLLGYIFVLTFF 285
G+ I T++ N++SF G+ ++R+FC S++ L +I +LT +
Sbjct 263 GMSILCTSINNILSFLTGTLLPIPALRSFCAQSSILLTFNFIAILTIY 310
> At1g42470
Length=1248
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 62/237 (26%), Positives = 109/237 (45%), Gaps = 30/237 (12%)
Query 70 TSASALMLHYDADG----RPMHEERNATWERLLIQVLK---------ANQTLGSLRAYFQ 116
+ ASA ++ Y D + E+ WE+ IQ+ K N TL
Sbjct 502 SEASAFLVTYPVDNFVDNKGNKTEKAVAWEKAFIQLAKDELLPMVQAKNLTLSFSSESSI 561
Query 117 AFRSRDDELKASTAESKDVVYVVFTFFLLASY-STALNFSCDLYR----SKLFSSLMGFF 171
+ + STA DV+ + ++ ++ +Y S L S L SK+ L G
Sbjct 562 EEELK----RESTA---DVITIAISYLVMFAYISLTLGDSPRLKSFYITSKVLLGLSGVL 614
Query 172 AAFMGLGAGMGIVCYMKVAMVPTVLI---CPFLVLGIGVDDVFVLLNAYCMGYTTHDPEQ 228
+ + +G + V M T++I PFLVL +GVD++ +L++A E+
Sbjct 615 LVMLSVLGSVGF--FSAVGMKSTLIIMEVIPFLVLAVGVDNMCILVHAVKRQEQELPLER 672
Query 229 RCIDTLKTSGLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMALLLGYIFVLTFF 285
R + L G IT+ +L +++F VG+F ++R F +++A+A+LL ++ +T F
Sbjct 673 RISNALMEVGPSITLASLAEILAFAVGAFIKMPAVRVFSMFAALAVLLDFLLQITAF 729
> CE00207
Length=413
Score = 56.6 bits (135), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 43/70 (61%), Gaps = 0/70 (0%)
Query 219 MGYTTHDPEQRCIDTLKTSGLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMALLLGY 278
M + + EQR I L S + +T+LT+ +SF +GS S + ++R FC Y AMA+L +
Sbjct 1 MTKKSSNEEQRYIHALTESAASLFLTSLTDGLSFAIGSISDFHAVRVFCTYCAMAILFMF 60
Query 279 IFVLTFFFPV 288
+F +TFF V
Sbjct 61 LFQVTFFNAV 70
> CE12664
Length=877
Score = 56.2 bits (134), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 33/93 (35%), Positives = 53/93 (56%), Gaps = 5/93 (5%)
Query 197 ICPFLVLGIGVDDVFVLLNAYCMGYTTHD----PEQRCIDTLKTSGLGITITTLTNLISF 252
+ PFL+L IGVDD ++ ++A CM T D ++ L G I IT++TNL +F
Sbjct 326 VTPFLILAIGVDDAYLQVHA-CMRLTAEDSSMTKREKIARMLVEVGPSIAITSMTNLFAF 384
Query 253 GVGSFSTYLSIRNFCVYSAMALLLGYIFVLTFF 285
VG ++ I FC +A+A+L +I+ +T +
Sbjct 385 LVGIYTPTPEISLFCAGNAVAILFDFIYQITMY 417
> CE29096
Length=1391
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 33/109 (30%), Positives = 60/109 (55%), Gaps = 3/109 (2%)
Query 180 GMGIVCYMKVAM-VPTVLICPFLVLGIGVDDVFVLLNAYCMGYTTHDPEQRCIDTLKTSG 238
G+G ++ + T + PFL LG+G+DD+F+LL+ Y + + + LK +G
Sbjct 698 GLGFATHLGINFNAATTQVVPFLSLGLGIDDMFLLLHNYDEIINICNKNEIGV-LLKETG 756
Query 239 LGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMALLLGYIFVLTFFFP 287
+ + +T++ N+++F G ++R+FC +A+ L IF L F FP
Sbjct 757 MSVMLTSINNILAFISGYVLPIPALRSFCSQTAILLAFNLIF-LMFIFP 804
> CE25194
Length=1388
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 33/109 (30%), Positives = 60/109 (55%), Gaps = 3/109 (2%)
Query 180 GMGIVCYMKVAM-VPTVLICPFLVLGIGVDDVFVLLNAYCMGYTTHDPEQRCIDTLKTSG 238
G+G ++ + T + PFL LG+G+DD+F+LL+ Y + + + LK +G
Sbjct 698 GLGFATHLGINFNAATTQVVPFLSLGLGIDDMFLLLHNYDEIINICNKNEIGV-LLKETG 756
Query 239 LGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMALLLGYIFVLTFFFP 287
+ + +T++ N+++F G ++R+FC +A+ L IF L F FP
Sbjct 757 MSVMLTSINNILAFISGYVLPIPALRSFCSQTAILLAFNLIF-LMFIFP 804
> At4g38350
Length=1054
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 52/89 (58%), Gaps = 0/89 (0%)
Query 197 ICPFLVLGIGVDDVFVLLNAYCMGYTTHDPEQRCIDTLKTSGLGITITTLTNLISFGVGS 256
+ PFLVL +GVD++ +L++A EQR L G IT+ +L+ +++F VG+
Sbjct 613 VIPFLVLAVGVDNMCILVHAVKRQPREVSLEQRISSALVEVGPSITLASLSEVLAFAVGA 672
Query 257 FSTYLSIRNFCVYSAMALLLGYIFVLTFF 285
F + R F +++A+A++L + +T F
Sbjct 673 FVPMPACRIFSMFAALAIMLDFFLQITAF 701
> Hs18598817
Length=781
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 54/228 (23%), Positives = 101/228 (44%), Gaps = 9/228 (3%)
Query 63 LGSIECITSASALML-HYDADGRPMHEERNATWERLLIQVLKANQTLGS-LRAY-FQAFR 119
+ S + + SA A+ L +Y +++ WE ++ Q S ++ Y + +
Sbjct 91 VHSKDRVKSAEAIQLTYYLQSINSLNDMVAERWESSFCDTVRLFQKSNSKVKMYPYTSSS 150
Query 120 SRDDELKASTAESKDVVYVVFTFFLLASYSTALNFSCDLYRSKLFSSLMGFFAAFMGLGA 179
R+D K S + Y+V + L+ + + D RSK + L+G +
Sbjct 151 LREDFQKTSRVSER---YLVTSLILVVTMAILCCSMQDCVRSKPWLGLLGLVTISLATLT 207
Query 180 GMGIVCYMKVAMVPTVLICPFLVLGIGVDDVFVLLNAYCMGYTTHDPEQRCIDTLKTSGL 239
GI+ T L PF V+G G+ F +L+++ ++R S L
Sbjct 208 AAGIINLTGGKYNSTFLGVPF-VIGHGLYGTFEMLSSWRKTREDQHVKERTAAVYADSML 266
Query 240 GITITTLTNLISFGVGS--FSTYLSIRNFCVYSAMALLLGYIFVLTFF 285
++TT L++FG+G+ F+ + R FC S +A+ Y++VL+F+
Sbjct 267 SFSLTTAMYLVTFGIGASPFTNIEAARIFCCNSCIAIFFNYLYVLSFY 314
> YPL006w
Length=1170
Score = 50.8 bits (120), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 48/206 (23%), Positives = 91/206 (44%), Gaps = 9/206 (4%)
Query 87 HEERNATWERLLIQVLKANQTLGSLRAYFQAFRSRDDELKASTAESKDVVYVVFTFFLLA 146
H + WE L + L + LR F S + EL + D+ V ++ ++
Sbjct 514 HTQSANRWEERLEEYLLDLKVPEGLRISFNTEISLEKELN----NNNDISTVAISYLMMF 569
Query 147 SYST-ALNFSCDLYRSKLFSSLMGFFAAFMGLGAGMGIVCYMKVAMVPTVLICPFLVLGI 205
Y+T AL R L S + A + AG + +K ++ +I PFL+L I
Sbjct 570 LYATWALRRKDGKTRLLLGISGLLIVLASIVCAAGFLTLFGLKSTLIIAEVI-PFLILAI 628
Query 206 GVDDVFVLLNAY---CMGYTTHDPEQRCIDTLKTSGLGITITTLTNLISFGVGSFSTYLS 262
G+D++F++ + Y C + +Q+ I + I ++ L F + +F T +
Sbjct 629 GIDNIFLITHEYDRNCEQKPEYSIDQKIISAIGRMSPSILMSLLCQTGCFLIAAFVTMPA 688
Query 263 IRNFCVYSAMALLLGYIFVLTFFFPV 288
+ NF +YS ++++ + LT + +
Sbjct 689 VHNFAIYSTVSVIFNGVLQLTAYVSI 714
> CE15305
Length=983
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 49/236 (20%), Positives = 97/236 (41%), Gaps = 14/236 (5%)
Query 63 LGSIECITSASALMLHYDADGRP-MHEERNATWERLLIQVLKANQTLGSLRAYFQAFRSR 121
+ I + S + L + A+ +P E + WE L+ + + L+ Y +
Sbjct 242 VSRITNVKSVKMITLQFRAEHKPGWTEAQVKKWEMSLVDIFEKRYNSKRLKIYAYSQSYV 301
Query 122 DDELKASTAESKDVVYVVFTFFLLASYSTALNFSCDLYRSKLFSSLMGFFAAFMGLGAGM 181
++E+ + V F L S + + +++ + ++ A L A
Sbjct 302 EEEMVRGGIIMIPYLVVGFAIMCLCSCVLVMIRALYMHQENGYKIILAIMACLTPLLACA 361
Query 182 GIVCYM---KVAMVPTVLICPFLVLGIGVDDVFVLLNAYCMGYTTHDPE---------QR 229
+ M V + + PFLVL IGVD +++++ + T H E R
Sbjct 362 TALALMFLCGVRFASILCVIPFLVLSIGVDSSYLMIHEW-QRVTKHMRETPRKKDSVGHR 420
Query 230 CIDTLKTSGLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMALLLGYIFVLTFF 285
+ + G I I+ LTN+ + VGSF++ I C + +++ +I+ +TF+
Sbjct 421 MSEVMAEVGPAILISCLTNMFADAVGSFTSSPEITLLCTGNMLSMWFAFIYQMTFY 476
> CE23854
Length=1110
Score = 45.4 bits (106), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 39/163 (23%), Positives = 76/163 (46%), Gaps = 15/163 (9%)
Query 137 YVVFTFFLLASYSTALNFSCDLYRSKL----FS-SLMGFFAAFMGLGAGMGIVCYMKVAM 191
+++ F ++A S+ F +Y ++ FS ++ FM G +G + + V
Sbjct 472 FLIVGFVIMAIVSSVTTFFSAVYMQQVSIHKFSLAIAACICPFMACGTALGALFFCGVRF 531
Query 192 VPTVLICPFLVLGIGVDDVFVLLNAY--------CMGYTTHDPEQRCIDTLKTSGLGITI 243
+ + PFLVL IGVDD +++++++ P R + L +G I I
Sbjct 532 GSILCVTPFLVLAIGVDDAYLMIHSWQRVTAERRKHPVANDSPGSRLSEVLVDTGPAILI 591
Query 244 TTLTNLISFGVGSFSTYLSIRNFCVYSAMA-LLLGYIFVLTFF 285
+ LTN+ + G F++ I + Y MA + +++ +TF+
Sbjct 592 SALTNIFADVAGCFTSSPEI-SLLGYGNMACIFCDFLYQITFY 633
> 7304020
Length=1286
Score = 41.2 bits (95), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 32/103 (31%), Positives = 52/103 (50%), Gaps = 9/103 (8%)
Query 172 AAFMGLGAGMGIVCYMKVAMVPTVLICPFLVLGIGVDDVFVLLNAYCMGYTTHDPEQRCI 231
AA +GL A +GIV + + PFL LG+GVD +F+L AY ++ EQ +
Sbjct 473 AAGLGLSALLGIVFN-----AASTQVVPFLALGLGVDHIFMLTAAYA---ESNRREQTKL 524
Query 232 DTLKTSGLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMAL 274
LK G I + + SF +F +++ FC+ +A+ +
Sbjct 525 -ILKKVGPSILFSACSTAGSFFAAAFIPVPALKVFCLQAAIVM 566
> CE29251
Length=1383
Score = 38.1 bits (87), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 43/212 (20%), Positives = 86/212 (40%), Gaps = 15/212 (7%)
Query 89 ERNATWERLLIQVLKANQTLGSLRAY-FQAFRSRDDELKASTAESKDVVYVVFTFFL-LA 146
++ WE+ ++ K + + F A RS DE++ + V + F +
Sbjct 583 QKAELWEKEFLKFCKEYREKSPKVIFSFMAERSITDEIENDAKDEIVTVVIALAFLIGYV 642
Query 147 SYSTALNFSCD------LYRSKLFSSLMGFFAAFMGLGAGMGIVCYMKVAMVPTVLICPF 200
++S F C+ L S++ ++ + GI + V L+ F
Sbjct 643 TFSLGRYFVCENQLWSILVHSRICLGMLSVIINLLSSFCSWGIFSMFGIHPVKNALVVQF 702
Query 201 LVLGI-GVDDVFVLLNAYC-----MGYTTHDPEQRCIDTLKTSGL-GITITTLTNLISFG 253
V+ + GV F+++ Y M Y + D + + + + ++L SF
Sbjct 703 FVVTLLGVCRTFMVVKYYAQQRVSMPYMSPDQCPEIVGMVMAGTMPAMFSSSLGCAFSFF 762
Query 254 VGSFSTYLSIRNFCVYSAMALLLGYIFVLTFF 285
+G F+ +IR FC+Y+ +A+L+ + T F
Sbjct 763 IGGFTDLPAIRTFCLYAGLAVLIDVVLHCTIF 794
> Hs17462432
Length=1278
Score = 37.7 bits (86), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 35/155 (22%), Positives = 70/155 (45%), Gaps = 5/155 (3%)
Query 134 DVVYVVFTFFLLASYSTALNFSCDLYRSKLFSSLMGFFAAFMGLGAGMGIVCYMKVAMVP 193
+++ +V T+ +L +Y D+ +SK +L L +G+ C + + P
Sbjct 284 ELIPLVTTYIILFAYIYFSTRKIDMVKSKWGLALAAVVTVLSSLLMSVGL-CTL-FGLTP 341
Query 194 TV---LICPFLVLGIGVDDVFVLLNAYCMGYTTHDPEQRCIDTLKTSGLGITITTLTNLI 250
T+ I P+LV+ IG+++V VL + + + R L + I T L
Sbjct 342 TLNGGEIFPYLVVVIGLENVLVLTKSVVSTPVDLEVKLRIAQGLSSESWSIMKNMATELG 401
Query 251 SFGVGSFSTYLSIRNFCVYSAMALLLGYIFVLTFF 285
+G F+ +I+ FC+++ + L+ + + FF
Sbjct 402 IILIGYFTLVPAIQEFCLFAVVGLVSDFFLQMLFF 436
> 7301039
Length=920
Score = 35.4 bits (80), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 29/132 (21%), Positives = 57/132 (43%), Gaps = 2/132 (1%)
Query 156 CDLYR--SKLFSSLMGFFAAFMGLGAGMGIVCYMKVAMVPTVLICPFLVLGIGVDDVFVL 213
C L+R SK + G F F I+ ++ + FL+L I + + L
Sbjct 126 CSLHRLGSKYVLGIAGLFTVFSSFIFTTAIIKFLGSDISELKDALFFLLLVIDLSNSGRL 185
Query 214 LNAYCMGYTTHDPEQRCIDTLKTSGLGITITTLTNLISFGVGSFSTYLSIRNFCVYSAMA 273
G + Q L+ G I++ T+ ++ GVG+ S + C+++ ++
Sbjct 186 AQLALSGSNQAEVTQNIARGLELLGPAISLDTIVEVLLVGVGTLSGVQRLEVLCMFAVLS 245
Query 274 LLLGYIFVLTFF 285
+L+ Y+ +TF+
Sbjct 246 VLVNYVVFMTFY 257
> At1g77460
Length=2110
Score = 30.0 bits (66), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 37/78 (47%), Gaps = 6/78 (7%)
Query 81 ADGRPMHEERNATWERLLIQVLKANQTLGSLRAYFQ---AFRSRDDELKASTAESKDVVY 137
ADG P +RNA+ R L Q+LK L+ Q A S D LK+ +S D
Sbjct 746 ADGSP-EGKRNAS--RALHQLLKNFPVCDVLKGSAQCRFAILSLVDSLKSIDVDSADAFN 802
Query 138 VVFTFFLLASYSTALNFS 155
++ LLA + +NFS
Sbjct 803 ILEVVALLAKTKSGVNFS 820
Lambda K H
0.328 0.141 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6410666326
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40