bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_0731_orf2
Length=173
Score E
Sequences producing significant alignments: (Bits) Value
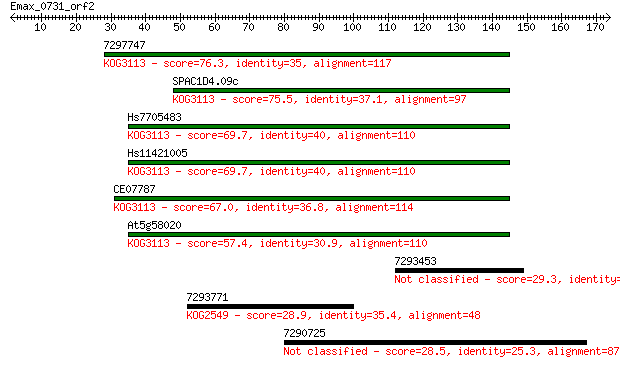
7297747 76.3 3e-14
SPAC1D4.09c 75.5 6e-14
Hs7705483 69.7 3e-12
Hs11421005 69.7 3e-12
CE07787 67.0 2e-11
At5g58020 57.4 1e-08
7293453 29.3 4.0
7293771 28.9 5.1
7290725 28.5 6.8
> 7297747
Length=299
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 41/132 (31%), Positives = 72/132 (54%), Gaps = 15/132 (11%)
Query 28 TQIREGDERLGKNESRDLRSSACAYSQEKLQPPLLACRIGRLYNKEAVIKALLEKA-LPP 86
++++ E+ K+ R+ R C +Q+ LQ P+ C +GRLY+K++VI+ LLEK +P
Sbjct 15 VRVKQKPEQKDKDAEREFRWRHCTLTQQSLQEPIAMCGMGRLYSKQSVIERLLEKEPMPE 74
Query 87 HMRHVKSLKDMKELR-------VEINASTGF-------PVCPITNADLSSGVRACIVWPC 132
HVKS+KD+++L E + + G +C + ++S R +W C
Sbjct 75 TAAHVKSMKDIRQLNPTPNPAFTEEDKTEGLLDTRHSPYICKLIGLEMSGKFRFVALWSC 134
Query 133 GFIISNRALEAM 144
G ++S RAL+ +
Sbjct 135 GCVLSERALKQI 146
> SPAC1D4.09c
Length=240
Score = 75.5 bits (184), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 36/99 (36%), Positives = 62/99 (62%), Gaps = 2/99 (2%)
Query 48 SACAYSQEKLQPPLLACRIGRLYNKEAVIKALLEK-ALPPHMRHVKSLKDMKELRVEINA 106
S CA + E L PP+++C +G+LYNK ++++ LL++ ++P H+KSLKD+ +L+VE++
Sbjct 38 SQCAITDEPLYPPIVSCGLGKLYNKASILQMLLDRSSVPKSPSHIKSLKDVVQLQVELDD 97
Query 107 STG-FPVCPITNADLSSGVRACIVWPCGFIISNRALEAM 144
S +CPIT +S + + PCG + AL+
Sbjct 98 SGKVLWLCPITRHVMSDTYQFAYIVPCGHVFEYSALKQF 136
> Hs7705483
Length=306
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 44/131 (33%), Positives = 70/131 (53%), Gaps = 21/131 (16%)
Query 35 ERLGKNESRDLRSSACAYSQEKLQPPLLACRIGRLYNKEAVIKALL----EKALPPHMRH 90
E++ K+ + + C SQE L+ P++AC +GRLYNK+AVI+ LL EKAL H
Sbjct 22 EKVDKDAELVAQWNYCTLSQEILRRPIVACELGRLYNKDAVIEFLLDKSAEKALGKAASH 81
Query 91 VKSLKDMKELRVEIN-ASTGFP----------------VCPITNADLSSGVRACIVWPCG 133
+KS+K++ EL++ N A G +CP+ +++ R C + CG
Sbjct 82 IKSIKNVTELKLSDNPAWEGDKGNTKGDKHDDLQRARFICPVVGLEMNGRHRFCFLRCCG 141
Query 134 FIISNRALEAM 144
+ S RAL+ +
Sbjct 142 CVFSERALKEI 152
> Hs11421005
Length=306
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 44/131 (33%), Positives = 70/131 (53%), Gaps = 21/131 (16%)
Query 35 ERLGKNESRDLRSSACAYSQEKLQPPLLACRIGRLYNKEAVIKALL----EKALPPHMRH 90
E++ K+ + + C SQE L+ P++AC +GRLYNK+AVI+ LL EKAL H
Sbjct 22 EKVDKDAELVAQWNYCTLSQEILRRPIVACELGRLYNKDAVIEFLLDKSAEKALGKAASH 81
Query 91 VKSLKDMKELRVEIN-ASTGFP----------------VCPITNADLSSGVRACIVWPCG 133
+KS+K++ EL++ N A G +CP+ +++ R C + CG
Sbjct 82 IKSIKNVTELKLSDNPAWEGDKGNTKGDKHDDLQRARFICPVVGLEMNGRHRFCFLRCCG 141
Query 134 FIISNRALEAM 144
+ S RAL+ +
Sbjct 142 CVFSERALKEI 152
> CE07787
Length=300
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 42/133 (31%), Positives = 69/133 (51%), Gaps = 21/133 (15%)
Query 31 REGDERLGKNESRDLRSSACAYSQEKLQPPLLACRIGRLYNKEAVIKALLEKAL--PPHM 88
R+ E+L K+ + C +Q L+ P++ACR G+LYNKE VI ++L K +
Sbjct 18 RKKKEKLDKHVKNATKWRNCQLTQLPLKRPVIACRFGKLYNKEDVINSILSKTISKSASA 77
Query 89 RHVKSLKDMKELRVEINA----------------STGFPVCPITNADLSSGVRACIV-WP 131
H+K KD EL++ +N T F +CPITN + +G+++ +V W
Sbjct 78 SHIKGPKDFVELKLTLNKDFKRGDVKGDDYNDVNQTEF-ICPITNVPM-NGIQSFLVNWQ 135
Query 132 CGFIISNRALEAM 144
CG + S +A + +
Sbjct 136 CGCVYSEKAQQEV 148
> At5g58020
Length=354
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 34/120 (28%), Positives = 60/120 (50%), Gaps = 11/120 (9%)
Query 35 ERLGKNESRDLRSSACAYSQEKLQPPLLACRIGRLYNKEAVIKALLEKALPPHMRHVKSL 94
+++ NE R + CA S E L P + +G L+NKE ++ ALL K LP ++K L
Sbjct 109 DKVDPNEQRLSKWLNCALSNEPLAEPCVIDLLGNLFNKEVLVHALLSKRLPKQFSYIKGL 168
Query 95 KDMKELRV----------EINASTGFPVCPITNADLSSGVRACIVWPCGFIISNRALEAM 144
KDM +++ + S F CP++ + + + + CG ++S +AL+ +
Sbjct 169 KDMVNIKLTPVAGSDGSSQDTTSAQFQ-CPVSGLEFNGKYKFFALRGCGHVMSAKALKEV 227
> 7293453
Length=307
Score = 29.3 bits (64), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 12/37 (32%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 112 VCPITNADLSSGVRACIVWPCGFIISNRALEAMTLKD 148
+CPIT+ LS+ V ++ P G +++ +E + KD
Sbjct 228 MCPITHDVLSNAVPCAVLRPTGDVVTMECVERLIRKD 264
> 7293771
Length=592
Score = 28.9 bits (63), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 28/50 (56%), Gaps = 3/50 (6%)
Query 52 YSQEKLQPPLLACRIGRLYNKEAV--IKALLEKALPPHMRHVKSLKDMKE 99
+ E+++P LL I +K A I+A+L+K PP +R ++S D E
Sbjct 379 FISERIEPHLLGTSISN-TDKTAAGHIRAMLQKCCPPILRQMRSAPDTAE 427
> 7290725
Length=2951
Score = 28.5 bits (62), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 22/87 (25%), Positives = 41/87 (47%), Gaps = 8/87 (9%)
Query 80 LEKALPPHMRHVKSLKDMKELRVEINASTGFPVCPITNADLSSGVRACIVWPCGFIISNR 139
++KA P + RHV+ L M++ I+ + +P N + + V+ P G I+ N+
Sbjct 326 MDKASPEYQRHVQHL--MEQPGEIISNTVEYPKP---NVKMITTVKRL---PDGTIVKNK 377
Query 140 ALEAMTLKDGESNKLDSNSNSSANNST 166
E L +S+ +N+ +N T
Sbjct 378 RYETEQLTPSQSHTTHKQTNNQTHNQT 404
Lambda K H
0.315 0.131 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2598880752
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40