bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
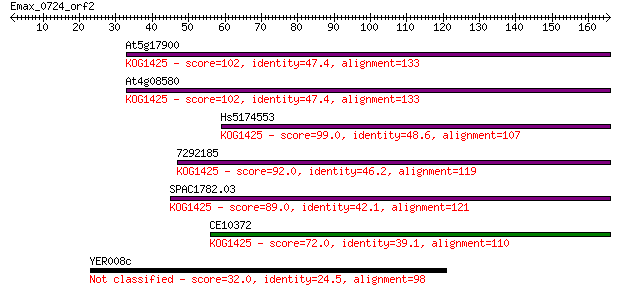
Query= Emax_0724_orf2
Length=165
Score E
Sequences producing significant alignments: (Bits) Value
At5g17900 102 4e-22
At4g08580 102 4e-22
Hs5174553 99.0 4e-21
7292185 92.0 5e-19
SPAC1782.03 89.0 4e-18
CE10372 72.0 5e-13
YER008c 32.0 0.59
> At5g17900
Length=435
Score = 102 bits (254), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 63/146 (43%), Positives = 94/146 (64%), Gaps = 20/146 (13%)
Query 33 QQNMLEGEEEMPD--DTDGLDPAQEYEDWKLRELERIRRDKEQLVAREKFL---EAVERR 87
++N+L E + D D L+ A+EYE WK RE+ RI+R+++ ARE L E +E+
Sbjct 237 RKNILLEEANIGDVETDDELNEAEEYEVWKTREIGRIKRERD---AREAMLREREEIEKL 293
Query 88 RQMTEEERREGDKELDK---MQPKREIKHKYQFMQKYYHRGAFFQ-----DLARSGEEPI 139
R MTE+ERR+ +++ K QPK+ K+ FMQKYYH+GAFFQ + +G + I
Sbjct 294 RNMTEQERRDWERKNPKPLSAQPKK----KWNFMQKYYHKGAFFQADPDDEAGSAGTDGI 349
Query 140 YLRDFNAPVGEDAVDKKALPKILQLR 165
+ RDF+AP GED +DK LPK++Q++
Sbjct 350 FQRDFSAPTGEDRLDKSILPKVMQVK 375
> At4g08580
Length=435
Score = 102 bits (253), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 63/146 (43%), Positives = 94/146 (64%), Gaps = 20/146 (13%)
Query 33 QQNMLEGEEEMPD--DTDGLDPAQEYEDWKLRELERIRRDKEQLVAREKFL---EAVERR 87
++N+L E + D D L+ A+EYE WK RE+ RI+R+++ ARE L E +E+
Sbjct 237 RKNILLEEANIGDVETDDELNEAEEYEVWKTREIGRIKRERD---AREAMLREREEIEKL 293
Query 88 RQMTEEERREGDKELDK---MQPKREIKHKYQFMQKYYHRGAFFQ-----DLARSGEEPI 139
R MTE+ERR+ +++ K QPK+ K+ FMQKYYH+GAFFQ + +G + I
Sbjct 294 RNMTEQERRDWERKNPKPSSAQPKK----KWNFMQKYYHKGAFFQADPDDEAGSAGTDGI 349
Query 140 YLRDFNAPVGEDAVDKKALPKILQLR 165
+ RDF+AP GED +DK LPK++Q++
Sbjct 350 FQRDFSAPTGEDRLDKSILPKVMQVK 375
> Hs5174553
Length=439
Score = 99.0 bits (245), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 52/107 (48%), Positives = 72/107 (67%), Gaps = 4/107 (3%)
Query 59 WKLRELERIRRDKEQLVAREKFLEAVERRRQMTEEERREGDKELDKMQPKREIKHKYQFM 118
WK+REL+RI+RD+E A EK +ER R +TEEERR + K+ + +K KY+F+
Sbjct 279 WKVRELKRIKRDREDREALEKEKAEIERMRNLTEEERRAELRANGKVITNKAVKGKYKFL 338
Query 119 QKYYHRGAFFQDLARSGEEPIYLRDFNAPVGEDAVDKKALPKILQLR 165
QKYYHRGAFF D +E +Y RDF+AP ED +K LPK++Q++
Sbjct 339 QKYYHRGAFFMD----EDEEVYKRDFSAPTLEDHFNKTILPKVMQVK 381
> 7292185
Length=478
Score = 92.0 bits (227), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 55/121 (45%), Positives = 76/121 (62%), Gaps = 8/121 (6%)
Query 47 TDGLDPAQEYEDWKLRELERIRRDKEQL--VAREKFLEAVERRRQMTEEERREGDKELDK 104
TD + EYE WKLREL+R++RD+E+ V REK ++R R MTEEERR+ ++ K
Sbjct 305 TDDENDEVEYEAWKLRELKRMKRDREERDNVEREKL--DIDRMRNMTEEERRQELRQNPK 362
Query 105 MQPKREIKHKYQFMQKYYHRGAFFQDLARSGEEPIYLRDFNAPVGEDAVDKKALPKILQL 164
+ + K KY+F+QKYYHRGAF+ D E + RDF ED DK LPK++Q+
Sbjct 363 VVTNKATKGKYKFLQKYYHRGAFYLD----EENDVLKRDFAQATLEDHFDKTILPKVMQV 418
Query 165 R 165
+
Sbjct 419 K 419
> SPAC1782.03
Length=355
Score = 89.0 bits (219), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 51/121 (42%), Positives = 74/121 (61%), Gaps = 5/121 (4%)
Query 45 DDTDGLDPAQEYEDWKLRELERIRRDKEQLVAREKFLEAVERRRQMTEEERREGDKELDK 104
DDTDG+DP EYE WKLR L R +RDKE+ + E+ A+E RR M EER D + +
Sbjct 183 DDTDGIDPQSEYELWKLRHLLRKKRDKEKSLELEREKMAIEERRLMNSEEREAQDLKDAE 242
Query 105 MQPKREIKHKYQFMQKYYHRGAFFQDLARSGEEPIYLRDFNAPVGEDAVDKKALPKILQL 164
+ + K QF+QKYYH+GAF+Q+ E+ + RD++ + ++K LPK +Q+
Sbjct 243 ASRRGKKKSSMQFLQKYYHKGAFYQN-----EDIVSKRDYSEATEGEVLNKDLLPKPMQI 297
Query 165 R 165
R
Sbjct 298 R 298
> CE10372
Length=466
Score = 72.0 bits (175), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 43/112 (38%), Positives = 71/112 (63%), Gaps = 8/112 (7%)
Query 56 YEDWKLRELERIRRDKEQL--VAREKFLEAVERRRQMTEEERREGDKELDKMQPKREIKH 113
YE WKLRE++R++R++++ AREK +++ M+EEER + + K+ ++ K
Sbjct 304 YEAWKLREMKRLKRNRDEREEAAREK--AELDKIHAMSEEERLKYLRLNPKVITNKQDKG 361
Query 114 KYQFMQKYYHRGAFFQDLARSGEEPIYLRDFNAPVGEDAVDKKALPKILQLR 165
KY+F+QKY+HRGAFF D E+ + R+F +D DK LPK++Q++
Sbjct 362 KYKFLQKYFHRGAFFLD----EEDEVLKRNFAEATNDDQFDKTILPKVMQVK 409
> YER008c
Length=1336
Score = 32.0 bits (71), Expect = 0.59, Method: Composition-based stats.
Identities = 24/98 (24%), Positives = 53/98 (54%), Gaps = 5/98 (5%)
Query 23 QQQQQQQQQQQQNMLEGEEEMPDDTDGLDPAQEYEDWKLRELERIRRDKEQLVAREKFLE 82
Q++ Q Q++ + LE E + + E++ R+LE R ++ + +K +E
Sbjct 348 QKRLQLQKENEMKRLEEERRIKQEERKRQMELEHQ----RQLEEEERKRQMELEAKKQME 403
Query 83 AVERRRQMTEEERREGDKELDKMQPKREIKHKYQFMQK 120
++R+RQ EE+R + ++EL ++Q K+ + + ++K
Sbjct 404 -LKRQRQFEEEQRLKKERELLEIQRKQREQETAERLKK 440
Lambda K H
0.314 0.131 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2353551590
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40